Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
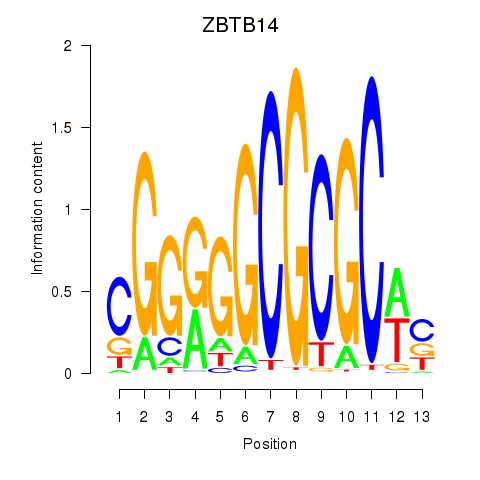
Results for ZBTB14
Z-value: 0.72
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.6 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5296138_5296194 | 0.70 | 1.2e-01 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_112451222 | 0.35 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr1_+_236305826 | 0.26 |
ENST00000366592.3
ENST00000366591.4 |
GPR137B
|
G protein-coupled receptor 137B |
| chr4_+_108852697 | 0.25 |
ENST00000508453.1
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr16_-_54963026 | 0.25 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr12_+_105724613 | 0.23 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr2_+_207308539 | 0.21 |
ENST00000374416.1
ENST00000374415.3 |
ADAM23
|
ADAM metallopeptidase domain 23 |
| chr2_+_136499287 | 0.20 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr11_-_126174186 | 0.19 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr18_-_45935663 | 0.19 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr1_+_53793885 | 0.19 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr22_-_31741757 | 0.18 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_125932685 | 0.18 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr19_-_14247365 | 0.17 |
ENST00000592798.1
ENST00000474890.1 |
ASF1B
|
anti-silencing function 1B histone chaperone |
| chr14_-_81902791 | 0.17 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr5_+_65440032 | 0.17 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr14_-_81902516 | 0.17 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr2_+_207308220 | 0.16 |
ENST00000264377.3
|
ADAM23
|
ADAM metallopeptidase domain 23 |
| chr9_-_35815013 | 0.16 |
ENST00000259667.5
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chr9_-_8857776 | 0.16 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr12_-_31479107 | 0.16 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr14_+_60716276 | 0.16 |
ENST00000528241.2
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr17_-_19648416 | 0.15 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_185826718 | 0.15 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr6_+_18155560 | 0.15 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chrX_-_107019181 | 0.15 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr4_-_153700864 | 0.15 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chrX_+_38420623 | 0.14 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr1_-_40367668 | 0.14 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr5_-_115177247 | 0.14 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr10_-_95209 | 0.14 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr1_+_111682827 | 0.14 |
ENST00000357172.4
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr3_+_194406603 | 0.13 |
ENST00000329759.4
|
FAM43A
|
family with sequence similarity 43, member A |
| chr18_-_5296138 | 0.13 |
ENST00000400143.3
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr1_+_40942887 | 0.13 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr19_+_44037546 | 0.13 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr2_+_149402009 | 0.13 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr15_+_73344791 | 0.13 |
ENST00000261908.6
|
NEO1
|
neogenin 1 |
| chr13_-_45151259 | 0.13 |
ENST00000493016.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr15_-_38856836 | 0.13 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr6_+_18155632 | 0.13 |
ENST00000297792.5
|
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr18_+_9102669 | 0.12 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr15_+_67813406 | 0.12 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr2_-_122042770 | 0.12 |
ENST00000263707.5
|
TFCP2L1
|
transcription factor CP2-like 1 |
| chr20_+_9049682 | 0.12 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr10_-_17659234 | 0.12 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr6_+_168227552 | 0.12 |
ENST00000400825.4
|
MLLT4
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
| chr11_-_134094420 | 0.12 |
ENST00000526422.1
ENST00000525485.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr5_-_178054105 | 0.12 |
ENST00000316308.4
|
CLK4
|
CDC-like kinase 4 |
| chr11_-_6440283 | 0.12 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr14_+_92789498 | 0.12 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_+_45613958 | 0.11 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr12_-_31744031 | 0.11 |
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr16_-_54962625 | 0.11 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr9_+_127020503 | 0.11 |
ENST00000545174.1
ENST00000444973.1 ENST00000454453.1 |
NEK6
|
NIMA-related kinase 6 |
| chr12_+_52345448 | 0.11 |
ENST00000257963.4
ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B
|
activin A receptor, type IB |
| chr2_+_203776937 | 0.11 |
ENST00000402905.3
ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF
|
calcium responsive transcription factor |
| chr9_+_6757634 | 0.11 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_+_111889212 | 0.11 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr9_+_6758024 | 0.11 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr8_+_97506033 | 0.11 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr2_-_40679148 | 0.11 |
ENST00000417271.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_-_168727713 | 0.11 |
ENST00000404867.3
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr14_+_60716159 | 0.11 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr15_-_71146347 | 0.11 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr4_-_1242463 | 0.11 |
ENST00000513420.1
|
CTBP1
|
C-terminal binding protein 1 |
| chr6_-_39197226 | 0.11 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr10_-_125651258 | 0.11 |
ENST00000241305.3
|
CPXM2
|
carboxypeptidase X (M14 family), member 2 |
| chr12_-_31743901 | 0.11 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr5_+_121647877 | 0.11 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr4_-_122618095 | 0.11 |
ENST00000515017.1
ENST00000501272.2 ENST00000296511.5 |
ANXA5
|
annexin A5 |
| chr15_-_71407806 | 0.11 |
ENST00000566432.1
ENST00000567117.1 |
CT62
|
cancer/testis antigen 62 |
| chr17_+_8924837 | 0.11 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr19_-_45004556 | 0.11 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr18_+_11103 | 0.11 |
ENST00000575820.1
ENST00000572573.1 |
AP005530.1
|
AP005530.1 |
| chr15_-_59041768 | 0.10 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr15_-_43882140 | 0.10 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr8_-_67525524 | 0.10 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr5_+_176560595 | 0.10 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr16_-_75498450 | 0.10 |
ENST00000566594.1
|
RP11-77K12.1
|
Uncharacterized protein |
| chr19_-_44809121 | 0.10 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr6_-_36355513 | 0.10 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr4_-_13549417 | 0.10 |
ENST00000501050.1
|
AC006445.8
|
long intergenic non-protein coding RNA 1096 |
| chr6_+_28227063 | 0.10 |
ENST00000343684.3
|
NKAPL
|
NFKB activating protein-like |
| chr8_-_42751820 | 0.10 |
ENST00000526349.1
ENST00000527424.1 ENST00000534961.1 ENST00000319073.4 |
RNF170
|
ring finger protein 170 |
| chr14_+_74058410 | 0.10 |
ENST00000326303.4
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr11_-_61348576 | 0.10 |
ENST00000263846.4
|
SYT7
|
synaptotagmin VII |
| chr18_-_51750948 | 0.10 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr3_-_185826286 | 0.10 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chr6_+_24495067 | 0.10 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr9_+_8858102 | 0.10 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr18_+_74534479 | 0.10 |
ENST00000320610.9
|
ZNF236
|
zinc finger protein 236 |
| chr9_-_114361919 | 0.10 |
ENST00000422125.1
|
PTGR1
|
prostaglandin reductase 1 |
| chr4_+_129731074 | 0.10 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr10_-_735553 | 0.09 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr2_+_29338236 | 0.09 |
ENST00000320081.5
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_+_72733350 | 0.09 |
ENST00000392613.5
ENST00000392612.3 ENST00000392610.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr9_+_71320596 | 0.09 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr5_-_168727786 | 0.09 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr1_-_84543614 | 0.09 |
ENST00000605506.1
|
RP11-486G15.2
|
RP11-486G15.2 |
| chr12_+_1100449 | 0.09 |
ENST00000360905.4
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr7_-_96654133 | 0.09 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr17_+_72428218 | 0.09 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr3_+_111578583 | 0.09 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr13_-_46626820 | 0.09 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr16_-_28223229 | 0.09 |
ENST00000566073.1
|
XPO6
|
exportin 6 |
| chr4_+_5053162 | 0.09 |
ENST00000282908.5
|
STK32B
|
serine/threonine kinase 32B |
| chr13_-_29069232 | 0.09 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr3_-_118753566 | 0.09 |
ENST00000491903.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr21_+_42688686 | 0.09 |
ENST00000398652.3
ENST00000398647.3 |
FAM3B
|
family with sequence similarity 3, member B |
| chrX_+_135579670 | 0.09 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr2_+_178077477 | 0.09 |
ENST00000411529.2
ENST00000435711.1 |
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr1_-_40367530 | 0.09 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr18_-_5296001 | 0.09 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr8_-_22014339 | 0.09 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr3_+_58477815 | 0.09 |
ENST00000404589.3
ENST00000490264.1 ENST00000491093.1 |
KCTD6
|
potassium channel tetramerization domain containing 6 |
| chr6_-_119256279 | 0.09 |
ENST00000316068.3
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr1_-_205719295 | 0.09 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr19_-_6110457 | 0.09 |
ENST00000586302.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr16_-_48419361 | 0.09 |
ENST00000394725.2
|
SIAH1
|
siah E3 ubiquitin protein ligase 1 |
| chr15_-_42783303 | 0.09 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr9_+_6758109 | 0.09 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr10_-_17659357 | 0.09 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr22_+_31742875 | 0.08 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr11_-_46142615 | 0.08 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_21279509 | 0.08 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr6_-_90121789 | 0.08 |
ENST00000359203.3
|
RRAGD
|
Ras-related GTP binding D |
| chr8_-_59572093 | 0.08 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr9_-_77703115 | 0.08 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr17_-_76837499 | 0.08 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr9_-_86571628 | 0.08 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chrX_+_146993648 | 0.08 |
ENST00000370470.1
|
FMR1
|
fragile X mental retardation 1 |
| chr5_+_176561129 | 0.08 |
ENST00000511258.1
ENST00000347982.4 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr7_-_158622157 | 0.08 |
ENST00000275418.7
|
ESYT2
|
extended synaptotagmin-like protein 2 |
| chr4_-_77134742 | 0.08 |
ENST00000452464.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr9_-_115095851 | 0.08 |
ENST00000343327.2
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr4_-_83934078 | 0.08 |
ENST00000505397.1
|
LIN54
|
lin-54 homolog (C. elegans) |
| chr2_-_128145498 | 0.08 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr16_-_1993124 | 0.08 |
ENST00000473663.1
ENST00000399753.2 ENST00000564908.1 |
MSRB1
|
methionine sulfoxide reductase B1 |
| chr19_-_13213662 | 0.08 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr2_-_203103185 | 0.08 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr1_+_182992545 | 0.08 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr2_-_75937994 | 0.08 |
ENST00000409857.3
ENST00000470503.1 ENST00000541687.1 ENST00000442309.1 |
GCFC2
|
GC-rich sequence DNA-binding factor 2 |
| chr1_+_210502238 | 0.08 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chr19_+_16435625 | 0.08 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr5_+_126112794 | 0.08 |
ENST00000261366.5
ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr12_+_112451120 | 0.08 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr11_-_47788985 | 0.08 |
ENST00000540172.2
|
FNBP4
|
formin binding protein 4 |
| chr1_-_246670614 | 0.08 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr10_+_105881779 | 0.08 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr6_+_127587755 | 0.08 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr1_+_36273743 | 0.08 |
ENST00000373210.3
|
AGO4
|
argonaute RISC catalytic component 4 |
| chr1_+_41249539 | 0.08 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr22_-_39096981 | 0.08 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr15_+_67814008 | 0.07 |
ENST00000557807.1
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr12_+_1100370 | 0.07 |
ENST00000543086.3
ENST00000546231.2 ENST00000397203.2 |
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr4_+_20255123 | 0.07 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr2_-_175547571 | 0.07 |
ENST00000409415.3
ENST00000359761.3 ENST00000272746.5 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr3_-_150481218 | 0.07 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr19_+_44037334 | 0.07 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr22_-_39548443 | 0.07 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr14_+_100070869 | 0.07 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr6_-_18155285 | 0.07 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr4_+_1873155 | 0.07 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr16_-_325910 | 0.07 |
ENST00000359740.5
ENST00000316163.5 ENST00000431291.2 ENST00000397770.3 ENST00000397768.3 |
RGS11
|
regulator of G-protein signaling 11 |
| chr6_+_31126291 | 0.07 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chrX_-_16888276 | 0.07 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr13_+_114238997 | 0.07 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr8_+_132916318 | 0.07 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr8_-_59572301 | 0.07 |
ENST00000038176.3
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr13_-_52027134 | 0.07 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr13_+_31774073 | 0.07 |
ENST00000343307.4
|
B3GALTL
|
beta 1,3-galactosyltransferase-like |
| chr6_+_24495185 | 0.07 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr11_+_65029421 | 0.07 |
ENST00000541089.1
|
POLA2
|
polymerase (DNA directed), alpha 2, accessory subunit |
| chr2_-_175869936 | 0.07 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr15_+_32322685 | 0.07 |
ENST00000454250.3
ENST00000306901.3 |
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr15_-_71407833 | 0.07 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr8_-_144679602 | 0.07 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr15_+_91643442 | 0.07 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr5_+_111755280 | 0.07 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr12_+_124457746 | 0.07 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr3_-_149688655 | 0.07 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr1_+_155051305 | 0.07 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr1_-_62784935 | 0.07 |
ENST00000354381.3
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr19_-_6110474 | 0.07 |
ENST00000587181.1
ENST00000587321.1 ENST00000586806.1 ENST00000589742.1 ENST00000592546.1 ENST00000303657.5 |
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr11_+_71791849 | 0.07 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr2_-_75938115 | 0.07 |
ENST00000321027.3
|
GCFC2
|
GC-rich sequence DNA-binding factor 2 |
| chr17_-_56065540 | 0.07 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_-_32906388 | 0.07 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr1_-_111682662 | 0.07 |
ENST00000286692.4
|
DRAM2
|
DNA-damage regulated autophagy modulator 2 |
| chr6_-_31080336 | 0.07 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chrX_-_153714917 | 0.07 |
ENST00000369653.4
|
UBL4A
|
ubiquitin-like 4A |
| chr8_+_87526732 | 0.07 |
ENST00000523469.1
ENST00000522240.1 |
CPNE3
|
copine III |
| chr17_+_56160768 | 0.07 |
ENST00000579991.2
|
DYNLL2
|
dynein, light chain, LC8-type 2 |
| chr9_-_100459639 | 0.07 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chrX_+_11776410 | 0.07 |
ENST00000361672.2
|
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr10_+_105036909 | 0.07 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr19_-_8070474 | 0.07 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr11_-_118023490 | 0.07 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr9_-_72287191 | 0.07 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr9_+_127020202 | 0.07 |
ENST00000373600.3
ENST00000320246.5 |
NEK6
|
NIMA-related kinase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.2 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.3 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.5 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0042424 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.0 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.0 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |