Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
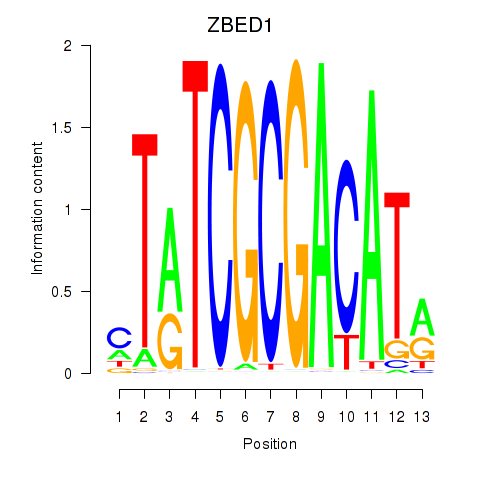
Results for ZBED1
Z-value: 0.35
Transcription factors associated with ZBED1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBED1
|
ENSG00000214717.5 | zinc finger BED-type containing 1 |
|
ZBED1
|
ENSGR0000214717.5 | zinc finger BED-type containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBED1 | hg19_v2_chrX_-_2418596_2418621 | 0.65 | 1.7e-01 | Click! |
Activity profile of ZBED1 motif
Sorted Z-values of ZBED1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_7760457 | 0.29 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr17_+_4843594 | 0.25 |
ENST00000570328.1
|
RNF167
|
ring finger protein 167 |
| chr17_-_4843316 | 0.22 |
ENST00000544061.2
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr17_-_7760779 | 0.18 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr19_+_54641444 | 0.16 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr17_+_4843352 | 0.14 |
ENST00000573404.1
ENST00000576452.1 |
RNF167
|
ring finger protein 167 |
| chr6_+_134273321 | 0.14 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chr17_-_7761172 | 0.14 |
ENST00000333775.5
ENST00000575771.1 |
LSMD1
|
LSM domain containing 1 |
| chr17_-_7761256 | 0.13 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr17_-_4843206 | 0.12 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr7_+_139025875 | 0.10 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr1_+_151254738 | 0.07 |
ENST00000336715.6
ENST00000324048.5 ENST00000368879.2 |
ZNF687
|
zinc finger protein 687 |
| chr1_+_36023370 | 0.07 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr22_+_19701985 | 0.06 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr2_+_74685413 | 0.06 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr17_+_4843303 | 0.05 |
ENST00000571816.1
|
RNF167
|
ring finger protein 167 |
| chr17_+_4843413 | 0.05 |
ENST00000572430.1
ENST00000262482.6 |
RNF167
|
ring finger protein 167 |
| chr2_-_677369 | 0.05 |
ENST00000281017.3
|
TMEM18
|
transmembrane protein 18 |
| chr17_+_4843679 | 0.04 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chrX_-_53711064 | 0.03 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr17_+_4843654 | 0.03 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167 |
| chr17_+_7761301 | 0.01 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr2_-_219906220 | 0.01 |
ENST00000458526.1
ENST00000409865.3 ENST00000410037.1 ENST00000457968.1 ENST00000436631.1 ENST00000341552.5 ENST00000441968.1 ENST00000295729.2 |
CCDC108
|
coiled-coil domain containing 108 |
| chr1_-_184723701 | 0.01 |
ENST00000367512.3
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr7_-_150924121 | 0.01 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr1_-_184723942 | 0.01 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBED1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |