Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
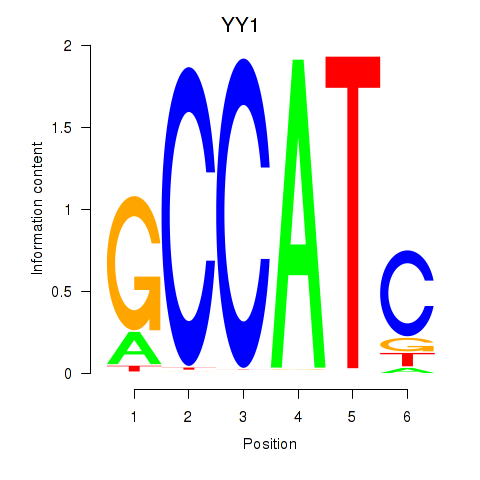
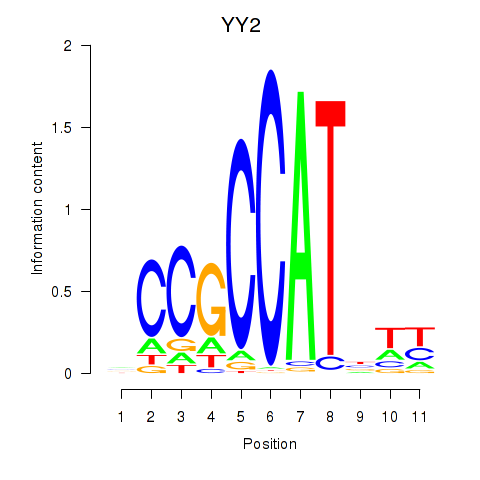
Results for YY1_YY2
Z-value: 1.13
Transcription factors associated with YY1_YY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
YY1
|
ENSG00000100811.6 | YY1 transcription factor |
|
YY2
|
ENSG00000230797.2 | YY2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| YY2 | hg19_v2_chrX_+_21874105_21874105 | 0.72 | 1.1e-01 | Click! |
| YY1 | hg19_v2_chr14_+_100705322_100705360 | -0.44 | 3.8e-01 | Click! |
Activity profile of YY1_YY2 motif
Sorted Z-values of YY1_YY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_153236819 | 1.33 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr19_-_41903161 | 1.19 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chrX_+_153237740 | 1.16 |
ENST00000369982.4
|
TMEM187
|
transmembrane protein 187 |
| chrX_-_153237258 | 1.02 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr4_+_76439095 | 0.95 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr9_-_35658007 | 0.92 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr19_+_797392 | 0.87 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chrX_-_153236620 | 0.83 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr19_+_18043810 | 0.78 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr19_-_13044494 | 0.77 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr6_-_27806117 | 0.76 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr15_+_42565393 | 0.71 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr17_-_47865948 | 0.65 |
ENST00000513602.1
|
FAM117A
|
family with sequence similarity 117, member A |
| chr19_+_55897699 | 0.64 |
ENST00000558131.1
ENST00000558752.1 ENST00000458349.2 |
RPL28
|
ribosomal protein L28 |
| chr19_+_797443 | 0.64 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr16_-_2827128 | 0.64 |
ENST00000494946.2
ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr11_-_47270341 | 0.62 |
ENST00000529444.1
ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr17_+_74732889 | 0.60 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr19_+_1383890 | 0.59 |
ENST00000539480.1
ENST00000313408.7 ENST00000414651.2 |
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chrX_+_153059608 | 0.58 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr19_-_19030157 | 0.57 |
ENST00000349893.4
ENST00000351079.4 ENST00000600932.1 ENST00000262812.4 |
COPE
|
coatomer protein complex, subunit epsilon |
| chr19_-_3985455 | 0.57 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr17_-_17399701 | 0.57 |
ENST00000225688.3
ENST00000579152.1 |
RASD1
|
RAS, dexamethasone-induced 1 |
| chr19_+_36545833 | 0.56 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr19_+_17326521 | 0.54 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr16_+_30662085 | 0.53 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr19_-_10946949 | 0.53 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr18_-_47814032 | 0.51 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr5_-_180671172 | 0.50 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr6_+_31588478 | 0.50 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr6_+_26020672 | 0.49 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr18_-_47813940 | 0.49 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr8_-_11058847 | 0.49 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr1_-_149814478 | 0.47 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr16_+_77233294 | 0.46 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_+_19030478 | 0.44 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr11_-_62477103 | 0.44 |
ENST00000532818.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_-_30006922 | 0.44 |
ENST00000564026.1
|
HIRIP3
|
HIRA interacting protein 3 |
| chr20_-_34542548 | 0.43 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr19_+_984313 | 0.43 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr19_-_39322299 | 0.43 |
ENST00000601094.1
ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr17_-_7760457 | 0.42 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr12_-_62653903 | 0.42 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr12_-_72057571 | 0.41 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr16_-_67260901 | 0.41 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr19_-_10946871 | 0.39 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr19_+_19030497 | 0.39 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr19_+_56111680 | 0.38 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr19_+_17830051 | 0.37 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr1_-_1167346 | 0.37 |
ENST00000545427.1
ENST00000360001.6 |
SDF4
|
stromal cell derived factor 4 |
| chr11_-_64889649 | 0.37 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr19_-_7694417 | 0.37 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr17_-_4843316 | 0.37 |
ENST00000544061.2
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chrX_-_153059958 | 0.36 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr20_+_44563267 | 0.36 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr16_+_2022036 | 0.36 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr3_+_47866490 | 0.36 |
ENST00000457607.1
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr19_+_17326141 | 0.36 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr6_-_27840099 | 0.36 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr19_+_4791722 | 0.35 |
ENST00000269856.3
|
FEM1A
|
fem-1 homolog a (C. elegans) |
| chrX_+_153060090 | 0.35 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr4_-_2010562 | 0.35 |
ENST00000411649.1
ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA
|
negative elongation factor complex member A |
| chr16_+_30709530 | 0.35 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_-_54518865 | 0.35 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59 |
| chr19_-_1848451 | 0.34 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr19_+_39421556 | 0.33 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr22_+_30752963 | 0.33 |
ENST00000445005.1
ENST00000430839.1 |
CCDC157
|
coiled-coil domain containing 157 |
| chr4_+_53525573 | 0.33 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr16_+_30662360 | 0.33 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr1_-_153950116 | 0.32 |
ENST00000368589.1
|
JTB
|
jumping translocation breakpoint |
| chr16_+_30662184 | 0.32 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr16_-_30773372 | 0.32 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr1_-_1167411 | 0.32 |
ENST00000263741.7
|
SDF4
|
stromal cell derived factor 4 |
| chr22_+_30752606 | 0.32 |
ENST00000399824.2
ENST00000405659.1 ENST00000338306.3 |
CCDC157
|
coiled-coil domain containing 157 |
| chr22_+_35796108 | 0.31 |
ENST00000382011.5
ENST00000416905.1 |
MCM5
|
minichromosome maintenance complex component 5 |
| chr17_-_42767092 | 0.31 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr11_-_111957451 | 0.31 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr8_-_145688231 | 0.31 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr1_+_16693578 | 0.31 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr6_+_75994755 | 0.31 |
ENST00000607799.1
|
RP1-234P15.4
|
RP1-234P15.4 |
| chr6_+_27775899 | 0.30 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr1_+_161129240 | 0.30 |
ENST00000492950.1
|
USP21
|
ubiquitin specific peptidase 21 |
| chrX_-_153059811 | 0.30 |
ENST00000427365.2
ENST00000444450.1 ENST00000370093.1 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr12_+_14927270 | 0.30 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr3_+_52489503 | 0.30 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr5_+_134210035 | 0.30 |
ENST00000506916.1
|
TXNDC15
|
thioredoxin domain containing 15 |
| chr2_+_113342011 | 0.30 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_+_64961026 | 0.29 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr16_+_2079501 | 0.29 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr6_-_27860956 | 0.29 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr11_-_62476694 | 0.29 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_153950098 | 0.29 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr22_-_50699972 | 0.29 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr11_-_104480019 | 0.29 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr16_+_2273645 | 0.29 |
ENST00000565413.1
|
E4F1
|
E4F transcription factor 1 |
| chr8_+_145515263 | 0.29 |
ENST00000528838.1
|
HSF1
|
heat shock transcription factor 1 |
| chr7_+_766850 | 0.29 |
ENST00000437419.1
ENST00000440747.1 |
HEATR2
|
HEAT repeat containing 2 |
| chr16_+_30006997 | 0.29 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr1_+_107599267 | 0.28 |
ENST00000361318.5
ENST00000370078.1 |
PRMT6
|
protein arginine methyltransferase 6 |
| chr19_+_9203855 | 0.28 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr6_-_26032288 | 0.28 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr11_-_73587719 | 0.28 |
ENST00000545127.1
ENST00000537289.1 ENST00000355693.4 |
COA4
|
cytochrome c oxidase assembly factor 4 homolog (S. cerevisiae) |
| chr19_+_55897297 | 0.28 |
ENST00000431533.2
ENST00000428193.2 ENST00000558815.1 ENST00000560583.1 ENST00000560055.1 ENST00000559463.1 |
RPL28
|
ribosomal protein L28 |
| chr19_-_13068012 | 0.28 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr11_+_47270436 | 0.28 |
ENST00000395397.3
ENST00000405576.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_+_128502895 | 0.28 |
ENST00000492758.1
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr19_+_10812108 | 0.27 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr16_+_2273558 | 0.27 |
ENST00000301727.4
ENST00000565090.1 ENST00000564139.1 |
E4F1
|
E4F transcription factor 1 |
| chr2_+_74757050 | 0.27 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr11_-_62477313 | 0.27 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr9_+_131873842 | 0.27 |
ENST00000417728.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_-_152623231 | 0.27 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_+_149402009 | 0.27 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_-_67260691 | 0.27 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr19_-_39322497 | 0.26 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr11_-_795286 | 0.26 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr11_+_842928 | 0.26 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr14_-_91282821 | 0.26 |
ENST00000553948.1
|
TTC7B
|
tetratricopeptide repeat domain 7B |
| chr20_-_62203808 | 0.26 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr5_-_140027357 | 0.26 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr16_-_1821496 | 0.26 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr14_-_89960395 | 0.26 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr14_+_97925151 | 0.26 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr16_-_88772670 | 0.26 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr19_-_19774473 | 0.26 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr17_+_30814707 | 0.26 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr11_+_65779283 | 0.25 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr20_-_44600810 | 0.25 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr20_-_62129163 | 0.25 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr12_-_50236907 | 0.25 |
ENST00000333924.4
|
BCDIN3D
|
BCDIN3 domain containing |
| chr11_+_842808 | 0.25 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr16_-_1020954 | 0.25 |
ENST00000543238.1
ENST00000539379.1 ENST00000399843.2 ENST00000262301.11 |
LMF1
|
lipase maturation factor 1 |
| chr16_+_30662050 | 0.25 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr9_-_131709858 | 0.25 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr3_+_122399697 | 0.25 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_+_1283639 | 0.25 |
ENST00000303400.4
ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA
|
macrophage erythroblast attacher |
| chr16_+_8736232 | 0.25 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr2_-_242576864 | 0.25 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr19_-_11039261 | 0.25 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr17_-_4843206 | 0.24 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr19_+_11546440 | 0.24 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr16_+_88772866 | 0.24 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr17_-_45918539 | 0.24 |
ENST00000584123.1
ENST00000578323.1 ENST00000407215.3 ENST00000290216.9 |
SCRN2
|
secernin 2 |
| chr17_+_42634844 | 0.24 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr19_-_36545649 | 0.24 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr19_+_11546153 | 0.24 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr17_-_3571934 | 0.24 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr1_-_143767881 | 0.24 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr16_+_1832902 | 0.24 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr11_-_62476965 | 0.24 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_-_102289611 | 0.23 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr1_+_36023370 | 0.23 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr1_+_1334914 | 0.23 |
ENST00000576232.1
ENST00000570344.1 |
RP4-758J18.2
|
HCG20425, isoform CRA_a; Uncharacterized protein; cDNA FLJ53815 |
| chr3_-_183966717 | 0.23 |
ENST00000446569.1
ENST00000418734.2 ENST00000397676.3 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr11_-_10830463 | 0.23 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_-_39661947 | 0.23 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr11_+_65686952 | 0.23 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr6_-_26271612 | 0.23 |
ENST00000305910.3
|
HIST1H3G
|
histone cluster 1, H3g |
| chr14_-_91976794 | 0.23 |
ENST00000555462.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr6_-_44281043 | 0.23 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr8_-_145515055 | 0.23 |
ENST00000526552.1
ENST00000529231.1 ENST00000307404.5 |
BOP1
|
block of proliferation 1 |
| chr1_-_149900122 | 0.23 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr16_-_25122735 | 0.23 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr22_+_50312316 | 0.23 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr16_-_57831914 | 0.23 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr7_-_72971934 | 0.23 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr16_+_30773636 | 0.22 |
ENST00000402121.3
ENST00000565995.1 ENST00000563683.1 ENST00000357890.5 ENST00000565931.1 |
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr10_-_75385711 | 0.22 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr19_+_17970693 | 0.22 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr7_+_766320 | 0.22 |
ENST00000297440.6
ENST00000313147.5 |
HEATR2
|
HEAT repeat containing 2 |
| chr2_-_25016251 | 0.22 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr11_-_18270182 | 0.22 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr19_-_11039188 | 0.22 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr19_+_58258202 | 0.22 |
ENST00000431353.1
|
ZNF776
|
zinc finger protein 776 |
| chr17_+_4843594 | 0.22 |
ENST00000570328.1
|
RNF167
|
ring finger protein 167 |
| chr9_+_131873659 | 0.22 |
ENST00000452489.2
ENST00000347048.4 ENST00000357197.4 ENST00000445241.1 ENST00000355007.3 ENST00000414331.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr9_+_131709966 | 0.22 |
ENST00000372577.2
|
NUP188
|
nucleoporin 188kDa |
| chr7_-_73097741 | 0.22 |
ENST00000395176.2
|
DNAJC30
|
DnaJ (Hsp40) homolog, subfamily C, member 30 |
| chr10_-_75532373 | 0.22 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr2_+_220306238 | 0.22 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr3_-_52090461 | 0.22 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr9_-_35689900 | 0.21 |
ENST00000378300.5
ENST00000329305.2 ENST00000360958.2 |
TPM2
|
tropomyosin 2 (beta) |
| chr19_+_532049 | 0.21 |
ENST00000606136.1
|
CDC34
|
cell division cycle 34 |
| chr2_-_152118276 | 0.21 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr10_+_104678032 | 0.21 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr19_-_39421377 | 0.21 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr5_+_131409476 | 0.21 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr22_+_51039098 | 0.21 |
ENST00000399912.1
ENST00000329492.3 ENST00000442429.2 ENST00000341339.4 |
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr16_-_75467274 | 0.21 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr7_+_76109827 | 0.21 |
ENST00000446820.2
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr17_-_7760779 | 0.21 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr5_-_180670880 | 0.21 |
ENST00000511566.1
ENST00000511900.1 ENST00000504726.1 ENST00000512968.1 ENST00000376817.4 ENST00000513027.1 ENST00000503081.1 ENST00000456394.2 |
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr11_+_5775923 | 0.21 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr15_-_41522889 | 0.21 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr20_+_43374421 | 0.21 |
ENST00000372861.3
|
KCNK15
|
potassium channel, subfamily K, member 15 |
| chr7_+_148982396 | 0.21 |
ENST00000418158.2
|
ZNF783
|
zinc finger family member 783 |
| chr11_-_842509 | 0.21 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr8_-_145911188 | 0.21 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr16_+_67927147 | 0.20 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr6_+_108616243 | 0.20 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of YY1_YY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 1.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 0.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.4 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 0.4 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.4 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.4 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.3 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.3 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.3 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.3 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.3 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.2 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.6 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 1.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.3 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.2 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.3 | GO:0051006 | positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) regulation of follicle-stimulating hormone secretion(GO:0046880) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.4 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1904597 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.0 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 1.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.0 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.0 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.0 | GO:0046666 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.0 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.0 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.1 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.0 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 1.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 3.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.5 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 0.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.5 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.1 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.6 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.2 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 1.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0060229 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 2.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.0 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |