Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
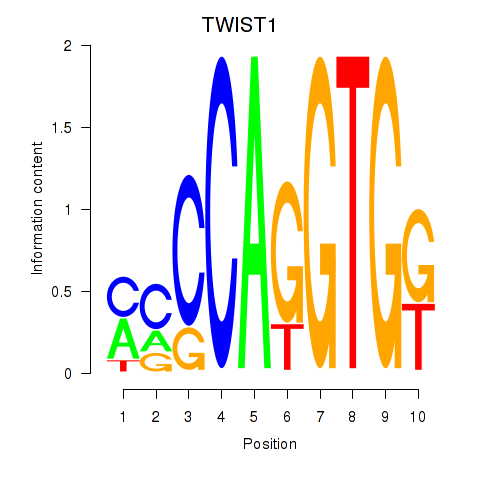
Results for TWIST1_SNAI1
Z-value: 0.85
Transcription factors associated with TWIST1_SNAI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TWIST1
|
ENSG00000122691.8 | twist family bHLH transcription factor 1 |
|
SNAI1
|
ENSG00000124216.3 | snail family transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SNAI1 | hg19_v2_chr20_+_48599506_48599536 | 0.27 | 6.1e-01 | Click! |
| TWIST1 | hg19_v2_chr7_-_19157248_19157295 | -0.20 | 7.0e-01 | Click! |
Activity profile of TWIST1_SNAI1 motif
Sorted Z-values of TWIST1_SNAI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150406527 | 0.79 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr19_-_18391708 | 0.69 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr1_+_27668505 | 0.63 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr7_+_149411860 | 0.59 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr2_+_121493717 | 0.59 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr2_+_220306238 | 0.55 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr7_-_120497178 | 0.47 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr15_+_40544749 | 0.45 |
ENST00000559617.1
ENST00000560684.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_219745020 | 0.44 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr1_-_95285652 | 0.42 |
ENST00000442418.1
|
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr17_-_39677971 | 0.39 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr1_-_1850697 | 0.39 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr19_-_56109119 | 0.39 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr19_+_18794470 | 0.38 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr19_+_55795493 | 0.36 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr19_-_48673465 | 0.35 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr9_-_140142222 | 0.34 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr12_+_65174519 | 0.33 |
ENST00000229088.6
|
TBC1D30
|
TBC1 domain family, member 30 |
| chr9_-_138987115 | 0.33 |
ENST00000277554.2
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing |
| chr17_+_79968655 | 0.33 |
ENST00000583744.1
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_+_1855645 | 0.32 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr1_-_16556038 | 0.32 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr17_+_43213004 | 0.32 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr9_+_92219919 | 0.31 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr19_-_5567842 | 0.29 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr19_-_6415695 | 0.29 |
ENST00000594496.1
ENST00000594745.1 ENST00000600480.1 |
KHSRP
|
KH-type splicing regulatory protein |
| chr1_+_60280458 | 0.28 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr14_-_23652849 | 0.28 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr4_-_11430221 | 0.28 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr21_+_37507210 | 0.27 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr19_-_10420459 | 0.27 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr5_-_42811986 | 0.27 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_+_35630926 | 0.26 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr17_+_4843594 | 0.26 |
ENST00000570328.1
|
RNF167
|
ring finger protein 167 |
| chr6_-_33160231 | 0.26 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr14_-_59932044 | 0.26 |
ENST00000395116.1
|
GPR135
|
G protein-coupled receptor 135 |
| chr20_+_43538692 | 0.26 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr4_+_142557717 | 0.26 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chrX_+_105969893 | 0.25 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr20_-_60942361 | 0.25 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr11_+_62379194 | 0.25 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr1_+_8378140 | 0.25 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr17_+_63096903 | 0.25 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr19_-_49843539 | 0.24 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr19_-_17932314 | 0.24 |
ENST00000598577.1
ENST00000317306.7 ENST00000379695.5 |
INSL3
|
insulin-like 3 (Leydig cell) |
| chr19_-_5567996 | 0.24 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr9_+_139746792 | 0.24 |
ENST00000317446.2
ENST00000445819.1 |
MAMDC4
|
MAM domain containing 4 |
| chr11_+_65779283 | 0.23 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr16_-_30393752 | 0.23 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr1_+_3385085 | 0.23 |
ENST00000445297.1
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr17_+_78193443 | 0.23 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr22_-_46373004 | 0.23 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr17_+_39421591 | 0.22 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr1_-_154178803 | 0.22 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr14_-_81902516 | 0.22 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr19_+_39897943 | 0.22 |
ENST00000600033.1
|
ZFP36
|
ZFP36 ring finger protein |
| chr14_+_77425972 | 0.22 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr8_-_53477968 | 0.21 |
ENST00000523939.1
ENST00000358543.4 |
FAM150A
|
family with sequence similarity 150, member A |
| chr16_-_850723 | 0.21 |
ENST00000248150.4
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr21_-_38338773 | 0.21 |
ENST00000399120.1
ENST00000419461.1 |
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr2_-_192016276 | 0.21 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr19_-_10628098 | 0.21 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr16_-_70712229 | 0.21 |
ENST00000562883.2
|
MTSS1L
|
metastasis suppressor 1-like |
| chr2_+_172864490 | 0.21 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr2_+_205410516 | 0.20 |
ENST00000406610.2
ENST00000462231.1 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr11_+_45168182 | 0.20 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr8_+_145726472 | 0.20 |
ENST00000528430.1
|
PPP1R16A
|
protein phosphatase 1, regulatory subunit 16A |
| chr6_-_75994025 | 0.20 |
ENST00000518161.1
|
TMEM30A
|
transmembrane protein 30A |
| chr11_-_2924720 | 0.20 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr16_+_3014269 | 0.19 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr20_+_43538756 | 0.19 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_+_67197288 | 0.19 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr20_+_49348109 | 0.19 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr1_-_85666688 | 0.19 |
ENST00000341460.5
|
SYDE2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
| chr7_+_74379083 | 0.19 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr19_+_41856816 | 0.18 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr17_-_71258491 | 0.18 |
ENST00000397671.1
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr13_+_103046954 | 0.18 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr4_-_168155169 | 0.18 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_7055631 | 0.18 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr8_+_22429205 | 0.18 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr19_-_33793430 | 0.18 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr6_+_1312675 | 0.18 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr1_+_33352036 | 0.18 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr5_+_76506706 | 0.18 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr14_+_77647966 | 0.18 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr4_+_106816644 | 0.17 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chr16_-_3086927 | 0.17 |
ENST00000572449.1
|
CCDC64B
|
coiled-coil domain containing 64B |
| chr6_-_30899924 | 0.17 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr7_+_24612848 | 0.17 |
ENST00000432190.1
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr10_+_99258625 | 0.17 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr15_+_31658349 | 0.17 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr2_+_30670077 | 0.17 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr16_+_718086 | 0.17 |
ENST00000315082.4
ENST00000563134.1 |
RHOT2
|
ras homolog family member T2 |
| chr19_-_12624647 | 0.17 |
ENST00000455490.1
|
ZNF709
|
zinc finger protein 709 |
| chr1_+_6508100 | 0.17 |
ENST00000461727.1
|
ESPN
|
espin |
| chr7_-_91875358 | 0.17 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr16_+_616995 | 0.17 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chr3_+_49059038 | 0.17 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr4_+_166300084 | 0.16 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr8_+_96037255 | 0.16 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_+_26274917 | 0.16 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr11_-_3013497 | 0.16 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr3_-_11761068 | 0.16 |
ENST00000418000.1
ENST00000417206.2 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr19_-_54676846 | 0.16 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr14_-_81902791 | 0.16 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr3_-_101395936 | 0.16 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr19_-_44008863 | 0.16 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr4_+_106816592 | 0.16 |
ENST00000379987.2
ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT
|
nephronectin |
| chr5_+_139027877 | 0.16 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr11_-_75141206 | 0.16 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr11_+_560956 | 0.16 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr14_+_23025534 | 0.15 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr9_+_19408919 | 0.15 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr19_-_54676884 | 0.15 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr8_+_21881636 | 0.15 |
ENST00000520125.1
ENST00000521157.1 ENST00000397940.1 ENST00000522813.1 |
NPM2
|
nucleophosmin/nucleoplasmin 2 |
| chr19_+_5904866 | 0.15 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr19_-_59010565 | 0.15 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr4_+_142557771 | 0.15 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr5_+_44809027 | 0.15 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr8_-_81083731 | 0.15 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr6_+_32811861 | 0.15 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr11_+_86511549 | 0.15 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr8_+_145490549 | 0.15 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr6_+_31540056 | 0.15 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr7_-_150780487 | 0.15 |
ENST00000482202.1
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr7_-_27169801 | 0.15 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr17_-_73761222 | 0.15 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr18_+_29078131 | 0.15 |
ENST00000585206.1
|
DSG2
|
desmoglein 2 |
| chr8_-_81083890 | 0.15 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr1_-_40254482 | 0.14 |
ENST00000397360.2
ENST00000372827.3 |
BMP8B
|
bone morphogenetic protein 8b |
| chr22_+_27068766 | 0.14 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr17_-_70417365 | 0.14 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr15_-_44969086 | 0.14 |
ENST00000434130.1
ENST00000560780.1 |
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr15_+_69365272 | 0.14 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr7_-_150780609 | 0.14 |
ENST00000297533.4
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr15_+_67547163 | 0.14 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr11_+_66824276 | 0.14 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr14_-_67982146 | 0.14 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr2_+_14772810 | 0.14 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr11_+_57509020 | 0.14 |
ENST00000388857.4
ENST00000528798.1 |
C11orf31
|
chromosome 11 open reading frame 31 |
| chr16_-_4664382 | 0.14 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr16_+_2106134 | 0.14 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr19_-_1490398 | 0.14 |
ENST00000588671.1
ENST00000300954.5 |
PCSK4
|
proprotein convertase subtilisin/kexin type 4 |
| chr11_+_706219 | 0.13 |
ENST00000533500.1
|
EPS8L2
|
EPS8-like 2 |
| chr12_-_62586543 | 0.13 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chrX_-_45710920 | 0.13 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr13_+_95364963 | 0.13 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr19_-_49565254 | 0.13 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr8_-_143823816 | 0.13 |
ENST00000246515.1
|
SLURP1
|
secreted LY6/PLAUR domain containing 1 |
| chr2_+_74881432 | 0.13 |
ENST00000453930.1
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr8_+_96037205 | 0.13 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr16_+_71660052 | 0.13 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr14_-_69262947 | 0.13 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr15_-_71055878 | 0.13 |
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr11_+_27062272 | 0.13 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_+_170288856 | 0.13 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr19_-_18717627 | 0.13 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr8_-_144815966 | 0.13 |
ENST00000388913.3
|
FAM83H
|
family with sequence similarity 83, member H |
| chr5_-_137071756 | 0.13 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr11_+_86511569 | 0.13 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr6_+_109416684 | 0.13 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr6_+_80341000 | 0.13 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr7_-_91875109 | 0.13 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr10_-_46970474 | 0.13 |
ENST00000503753.1
ENST00000374321.4 |
SYT15
|
synaptotagmin XV |
| chr17_-_73844722 | 0.13 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr14_-_69262789 | 0.13 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_223537475 | 0.12 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr5_+_94890840 | 0.12 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr7_+_2687173 | 0.12 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr6_+_31950150 | 0.12 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr12_-_121972556 | 0.12 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr8_-_145753940 | 0.12 |
ENST00000532827.1
|
C8orf82
|
chromosome 8 open reading frame 82 |
| chr2_+_220306745 | 0.12 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr8_-_99306564 | 0.12 |
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr3_+_42544084 | 0.12 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr15_+_81071684 | 0.12 |
ENST00000220244.3
ENST00000394685.3 ENST00000356249.5 |
KIAA1199
|
KIAA1199 |
| chr12_-_53601055 | 0.12 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr12_-_132628847 | 0.12 |
ENST00000397333.3
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr15_-_71055769 | 0.12 |
ENST00000539319.1
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr8_+_67687773 | 0.12 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr4_-_2420335 | 0.12 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr1_-_22215192 | 0.12 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr1_+_178694300 | 0.12 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_-_177116772 | 0.12 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr11_+_27062860 | 0.12 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_+_3708338 | 0.12 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr19_+_7710774 | 0.12 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chr5_+_7373230 | 0.12 |
ENST00000500616.2
|
CTD-2296D1.5
|
CTD-2296D1.5 |
| chr2_+_120517174 | 0.12 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr2_-_228028829 | 0.12 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr8_-_12612962 | 0.11 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr2_+_17721920 | 0.11 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr2_-_169746878 | 0.11 |
ENST00000282074.2
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr8_+_107670064 | 0.11 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr12_-_46663734 | 0.11 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr16_+_30675654 | 0.11 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr2_+_66666432 | 0.11 |
ENST00000495021.2
|
MEIS1
|
Meis homeobox 1 |
| chr19_+_2249308 | 0.11 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr20_+_9049742 | 0.11 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TWIST1_SNAI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 0.5 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.2 | 0.5 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.1 | 0.8 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.5 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 0.2 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.1 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 0.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.3 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:1902232 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) transformation of host cell by virus(GO:0019087) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.0 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.0 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.6 | GO:0060042 | retina morphogenesis in camera-type eye(GO:0060042) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0035768 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.0 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.1 | GO:0031300 | intrinsic component of organelle membrane(GO:0031300) |
| 0.1 | 0.3 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.3 | GO:0034678 | smooth muscle contractile fiber(GO:0030485) integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.2 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.2 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |