Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
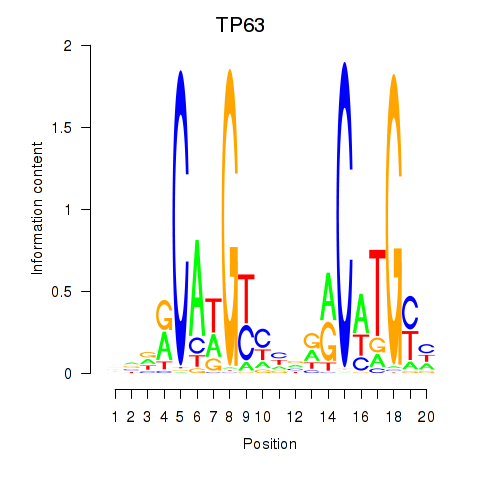
Results for TP63
Z-value: 1.04
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189507523_189507590 | 0.39 | 4.4e-01 | Click! |
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_3189861 | 0.84 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chrX_+_9880412 | 0.57 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr11_+_393428 | 0.55 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr16_+_57653596 | 0.51 |
ENST00000568791.1
ENST00000570044.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_17575584 | 0.44 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr16_+_57653625 | 0.42 |
ENST00000567553.1
ENST00000565314.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr6_-_33663474 | 0.40 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr9_-_88874519 | 0.40 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr7_+_79763271 | 0.38 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr16_+_57653989 | 0.38 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_-_4717835 | 0.37 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr16_+_1832902 | 0.37 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr5_-_42811986 | 0.36 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_+_80267973 | 0.35 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr9_+_35906176 | 0.35 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chrX_+_9880590 | 0.34 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr1_-_117753540 | 0.34 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr2_+_217524323 | 0.34 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr15_+_40532723 | 0.33 |
ENST00000558878.1
ENST00000558183.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chrX_-_138724677 | 0.32 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr17_+_1646130 | 0.32 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr16_+_67197288 | 0.31 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr17_+_78397320 | 0.31 |
ENST00000521634.1
ENST00000572886.1 |
ENDOV
|
endonuclease V |
| chr1_-_19229218 | 0.31 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr16_+_57653854 | 0.31 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_-_75467274 | 0.30 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr1_-_1356719 | 0.30 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr19_-_10420459 | 0.29 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr1_+_86889769 | 0.29 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr11_+_27062272 | 0.29 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_-_72439997 | 0.29 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr19_-_3500635 | 0.29 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr4_+_105828492 | 0.29 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr15_-_42448788 | 0.28 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr16_-_30006922 | 0.27 |
ENST00000564026.1
|
HIRIP3
|
HIRA interacting protein 3 |
| chr7_-_110174754 | 0.26 |
ENST00000435466.1
|
AC003088.1
|
AC003088.1 |
| chr19_+_507299 | 0.26 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr6_-_109330702 | 0.26 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr22_+_50628999 | 0.25 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr1_-_1356628 | 0.25 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr5_-_42812143 | 0.25 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_+_22960426 | 0.25 |
ENST00000540813.1
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
| chr11_-_77899634 | 0.24 |
ENST00000526208.1
ENST00000529350.1 ENST00000530018.1 ENST00000528776.1 |
KCTD21
|
potassium channel tetramerization domain containing 21 |
| chr11_-_111782484 | 0.24 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_111782696 | 0.24 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr1_-_38412683 | 0.23 |
ENST00000373024.3
ENST00000373023.2 |
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr7_-_72742085 | 0.23 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr9_-_119449483 | 0.23 |
ENST00000288520.5
ENST00000358637.4 ENST00000341734.4 |
ASTN2
|
astrotactin 2 |
| chr11_+_1856034 | 0.23 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr14_+_35514323 | 0.23 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chrX_-_111923145 | 0.23 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr4_+_671711 | 0.22 |
ENST00000400159.2
|
MYL5
|
myosin, light chain 5, regulatory |
| chr2_+_198365122 | 0.22 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr15_+_69365272 | 0.22 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr1_+_3607228 | 0.21 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr11_+_832804 | 0.21 |
ENST00000397420.3
ENST00000525718.1 |
CD151
|
CD151 molecule (Raph blood group) |
| chr4_+_105828537 | 0.21 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr12_-_122712038 | 0.21 |
ENST00000413918.1
ENST00000443649.3 |
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr1_-_19229014 | 0.21 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr15_-_72767490 | 0.21 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr16_-_70729496 | 0.20 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr3_-_49395892 | 0.20 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr15_+_42697065 | 0.20 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr8_+_117778736 | 0.20 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr17_+_38337491 | 0.20 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr6_+_31691121 | 0.20 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr1_+_223354486 | 0.19 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr2_+_208104497 | 0.19 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr7_-_100287071 | 0.19 |
ENST00000275732.5
|
GIGYF1
|
GRB10 interacting GYF protein 1 |
| chr1_+_38158090 | 0.19 |
ENST00000373055.1
ENST00000327331.2 |
CDCA8
|
cell division cycle associated 8 |
| chrX_-_129244336 | 0.19 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr3_-_33700589 | 0.19 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_76493294 | 0.19 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr1_-_19229248 | 0.19 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr11_+_18433840 | 0.18 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr22_-_38577782 | 0.18 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr2_+_169312725 | 0.18 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr19_-_11688260 | 0.18 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr20_-_34542548 | 0.18 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr4_-_40516560 | 0.18 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_-_180235755 | 0.18 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr19_-_19887185 | 0.17 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr20_+_57264187 | 0.17 |
ENST00000525967.1
ENST00000525817.1 |
NPEPL1
|
aminopeptidase-like 1 |
| chr19_+_15783879 | 0.17 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chr19_-_51504411 | 0.17 |
ENST00000593490.1
|
KLK8
|
kallikrein-related peptidase 8 |
| chr3_-_49395705 | 0.17 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr17_-_18430160 | 0.17 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr2_+_169312350 | 0.17 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr19_+_42364313 | 0.16 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr16_+_53483983 | 0.16 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr11_-_104972158 | 0.16 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_-_10747802 | 0.16 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr11_-_71814422 | 0.16 |
ENST00000278671.5
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr8_-_37351344 | 0.16 |
ENST00000520422.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr1_-_118727781 | 0.16 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr17_-_4938712 | 0.16 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr6_+_127898312 | 0.16 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr11_+_61583968 | 0.15 |
ENST00000517839.1
|
FADS2
|
fatty acid desaturase 2 |
| chr14_-_100046444 | 0.15 |
ENST00000554996.1
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr19_+_41094612 | 0.15 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr19_+_18496957 | 0.15 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr11_-_71814276 | 0.15 |
ENST00000538404.1
ENST00000535107.1 ENST00000545249.1 |
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr10_-_90751038 | 0.15 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr4_+_95916947 | 0.15 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_145218673 | 0.15 |
ENST00000326134.5
|
MROH1
|
maestro heat-like repeat family member 1 |
| chrX_-_138724994 | 0.15 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr18_-_19283649 | 0.15 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr1_+_24646263 | 0.15 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chrX_-_153602991 | 0.15 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr12_-_55042140 | 0.14 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr5_+_162864575 | 0.14 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr15_-_89456630 | 0.14 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr11_+_65686728 | 0.14 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr12_-_10321754 | 0.14 |
ENST00000539518.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr19_+_42364460 | 0.14 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chrX_+_153029633 | 0.14 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chrX_+_2670153 | 0.14 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr10_-_88729200 | 0.14 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr1_+_85527987 | 0.13 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr19_+_46498704 | 0.13 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr15_-_89456593 | 0.13 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr6_-_33385902 | 0.13 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr16_+_30006997 | 0.13 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr16_+_30007524 | 0.13 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr4_+_95917383 | 0.13 |
ENST00000512312.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr19_-_47231216 | 0.13 |
ENST00000594287.2
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr2_-_176033066 | 0.13 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr4_+_3388057 | 0.13 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr8_+_22437664 | 0.13 |
ENST00000436754.1
ENST00000426493.1 ENST00000429812.1 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr9_+_71944241 | 0.13 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr1_-_26197744 | 0.12 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr4_-_10023095 | 0.12 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chrX_-_153583257 | 0.12 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr4_-_129209221 | 0.12 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr6_+_35265586 | 0.12 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr22_+_40573921 | 0.12 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr17_-_34079897 | 0.12 |
ENST00000254466.6
ENST00000587565.1 |
GAS2L2
|
growth arrest-specific 2 like 2 |
| chr18_-_28742813 | 0.12 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr13_-_96296944 | 0.12 |
ENST00000361396.2
ENST00000376829.2 |
DZIP1
|
DAZ interacting zinc finger protein 1 |
| chr12_-_15942503 | 0.12 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr10_-_105212141 | 0.12 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr18_+_57567180 | 0.12 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr20_-_52790512 | 0.12 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr22_-_50946113 | 0.12 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr1_+_24645865 | 0.12 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr8_+_67025516 | 0.11 |
ENST00000517961.2
|
AC084082.3
|
AC084082.3 |
| chr19_-_5293243 | 0.11 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr11_-_93271058 | 0.11 |
ENST00000527149.1
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr5_-_64920115 | 0.11 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr1_+_24645807 | 0.11 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr10_-_105212059 | 0.11 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr3_+_69915363 | 0.11 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_27603582 | 0.11 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr3_+_152552685 | 0.11 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr15_-_90233866 | 0.11 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr22_-_38577731 | 0.11 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr1_+_203256898 | 0.11 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chr1_+_97187318 | 0.11 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr16_+_30006615 | 0.11 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr17_-_39459103 | 0.11 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr22_+_24999114 | 0.11 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr6_-_111804905 | 0.11 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr19_-_12662240 | 0.11 |
ENST00000416136.1
ENST00000428311.1 |
ZNF564
ZNF709
|
zinc finger protein 564 ZNF709 |
| chr1_+_24646002 | 0.11 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr6_+_148663729 | 0.10 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr8_+_22437965 | 0.10 |
ENST00000409141.1
ENST00000265810.4 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_17906970 | 0.10 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr12_+_3186521 | 0.10 |
ENST00000537971.1
ENST00000011898.5 |
TSPAN9
|
tetraspanin 9 |
| chr8_-_18942240 | 0.10 |
ENST00000521475.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_24645915 | 0.10 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr22_+_39378346 | 0.10 |
ENST00000407298.3
|
APOBEC3B
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
| chr19_-_11639931 | 0.10 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr3_+_23986748 | 0.10 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_+_67776012 | 0.10 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr8_-_67341208 | 0.10 |
ENST00000499642.1
|
RP11-346I3.4
|
RP11-346I3.4 |
| chr19_-_11688500 | 0.10 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr13_+_42846272 | 0.10 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr15_+_40532058 | 0.10 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr5_-_94417562 | 0.09 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_+_93558069 | 0.09 |
ENST00000371627.4
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr14_-_94595993 | 0.09 |
ENST00000238609.3
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2 |
| chr11_-_68611721 | 0.09 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr11_-_64889649 | 0.09 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr8_-_23021533 | 0.09 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr11_-_72432950 | 0.09 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_-_88974236 | 0.09 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr5_+_115420688 | 0.09 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr5_+_174905532 | 0.09 |
ENST00000502393.1
ENST00000506963.1 |
SFXN1
|
sideroflexin 1 |
| chr7_+_48211048 | 0.09 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr6_+_80129989 | 0.09 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr7_+_140396756 | 0.09 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chrX_-_45710920 | 0.09 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr2_+_112813134 | 0.08 |
ENST00000452614.1
|
TMEM87B
|
transmembrane protein 87B |
| chr15_+_41136263 | 0.08 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr19_-_46288917 | 0.08 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr17_+_79859985 | 0.08 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr1_-_46089718 | 0.08 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr2_+_178257372 | 0.08 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr20_-_43743790 | 0.08 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr17_+_20483037 | 0.08 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 0.7 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.3 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.3 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.3 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.4 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.4 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |