Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
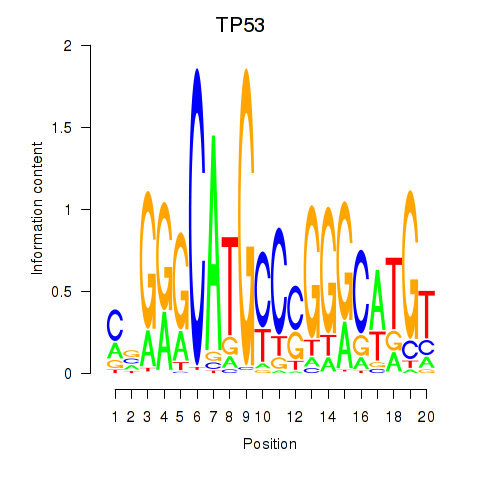
Results for TP53
Z-value: 0.70
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | tumor protein p53 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP53 | hg19_v2_chr17_-_7590745_7590856 | 0.66 | 1.6e-01 | Click! |
Activity profile of TP53 motif
Sorted Z-values of TP53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_26859300 | 0.61 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr18_+_44497455 | 0.42 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr2_+_197504278 | 0.42 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr4_+_186347388 | 0.36 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr1_-_54483803 | 0.31 |
ENST00000371360.1
ENST00000545928.1 |
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1 |
| chr22_-_24093267 | 0.29 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr11_+_65222698 | 0.28 |
ENST00000309775.7
|
AP000769.1
|
Uncharacterized protein |
| chr4_+_6784358 | 0.26 |
ENST00000508423.1
|
KIAA0232
|
KIAA0232 |
| chr3_+_32148106 | 0.25 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr1_+_54519242 | 0.23 |
ENST00000234827.1
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr22_+_29168652 | 0.23 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr6_-_109804412 | 0.23 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chrX_+_3189861 | 0.23 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chr1_-_54483842 | 0.22 |
ENST00000371362.3
ENST00000420619.1 |
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1 |
| chr19_+_28284803 | 0.21 |
ENST00000586220.1
ENST00000588784.1 ENST00000591549.1 ENST00000585827.1 ENST00000588636.1 ENST00000587188.1 |
CTC-459F4.3
|
CTC-459F4.3 |
| chr5_+_6766004 | 0.21 |
ENST00000506093.1
|
RP11-332J15.3
|
RP11-332J15.3 |
| chr15_+_100347228 | 0.21 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr2_+_11295624 | 0.21 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr19_-_14048804 | 0.21 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chr7_-_100239132 | 0.21 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr7_-_21985489 | 0.20 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr5_-_138775177 | 0.20 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr12_+_56546363 | 0.20 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr3_+_49977440 | 0.20 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr12_-_126467906 | 0.20 |
ENST00000507313.1
ENST00000545784.1 |
LINC00939
|
long intergenic non-protein coding RNA 939 |
| chr4_-_87374330 | 0.18 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_-_14788526 | 0.18 |
ENST00000438167.3
|
PLA2G10
|
phospholipase A2, group X |
| chrX_-_106959631 | 0.18 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_-_6591113 | 0.17 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr17_+_78234625 | 0.17 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr3_+_49977490 | 0.17 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr19_-_38714847 | 0.17 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr11_-_68611721 | 0.16 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr10_-_105212141 | 0.16 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr7_+_72742178 | 0.16 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr7_+_63361201 | 0.16 |
ENST00000450544.1
|
RP11-340I6.8
|
RP11-340I6.8 |
| chr3_+_49977523 | 0.15 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chrX_-_48827976 | 0.15 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr5_-_64920115 | 0.15 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr16_+_77756399 | 0.15 |
ENST00000564085.1
ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr8_-_49533521 | 0.15 |
ENST00000523038.1
|
RP11-567J20.1
|
RP11-567J20.1 |
| chr16_-_15149828 | 0.15 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr8_-_12293852 | 0.15 |
ENST00000262365.4
ENST00000351291.4 ENST00000309608.5 ENST00000527331.1 ENST00000532480.1 ENST00000393715.3 |
FAM86B2
|
family with sequence similarity 86, member B2 |
| chr6_+_139135648 | 0.14 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr19_+_57874835 | 0.14 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr4_+_6784401 | 0.13 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr16_-_15149917 | 0.13 |
ENST00000287706.3
|
NTAN1
|
N-terminal asparagine amidase |
| chr19_+_51293672 | 0.13 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr4_-_129209221 | 0.13 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr2_+_198365095 | 0.13 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr4_+_103422499 | 0.13 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr1_+_204494618 | 0.12 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr1_+_2477831 | 0.12 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr10_+_96305535 | 0.12 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chr5_-_168006324 | 0.12 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr5_-_72861175 | 0.12 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr1_-_169396666 | 0.12 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr11_-_47546250 | 0.12 |
ENST00000543178.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr13_+_38923959 | 0.12 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr18_-_28622699 | 0.11 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr15_+_79165296 | 0.11 |
ENST00000558746.1
ENST00000558830.1 ENST00000559345.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr22_+_29279552 | 0.11 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr8_-_29592736 | 0.11 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr15_+_79165372 | 0.11 |
ENST00000558502.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr18_-_28622774 | 0.11 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr11_-_77122928 | 0.11 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_-_105212059 | 0.11 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr7_+_72742162 | 0.11 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr15_+_79165151 | 0.10 |
ENST00000331268.5
|
MORF4L1
|
mortality factor 4 like 1 |
| chr2_+_208104497 | 0.10 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr10_+_90750493 | 0.10 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr14_+_36295638 | 0.10 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr5_+_72861560 | 0.10 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr1_-_114447620 | 0.10 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr7_-_100493744 | 0.10 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr1_+_211432700 | 0.10 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr7_-_74221288 | 0.10 |
ENST00000451013.2
|
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr15_+_79165222 | 0.10 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr19_-_28284793 | 0.10 |
ENST00000590523.1
|
LINC00662
|
long intergenic non-protein coding RNA 662 |
| chr4_+_153457404 | 0.09 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr6_-_114292284 | 0.09 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr5_+_178487354 | 0.09 |
ENST00000315475.6
|
ZNF354C
|
zinc finger protein 354C |
| chr22_-_28392227 | 0.09 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr15_-_63448973 | 0.09 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr14_-_64971288 | 0.09 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr19_+_42746927 | 0.09 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr9_-_116037840 | 0.09 |
ENST00000374206.3
|
CDC26
|
cell division cycle 26 |
| chr3_+_183894737 | 0.09 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr15_+_79165112 | 0.09 |
ENST00000426013.2
|
MORF4L1
|
mortality factor 4 like 1 |
| chr12_+_44229846 | 0.09 |
ENST00000551577.1
ENST00000266534.3 |
TMEM117
|
transmembrane protein 117 |
| chr2_-_18741882 | 0.09 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr7_+_116165754 | 0.09 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr11_+_5712234 | 0.09 |
ENST00000414641.1
|
TRIM22
|
tripartite motif containing 22 |
| chr2_+_177053307 | 0.09 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr14_-_36789865 | 0.09 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr1_-_114447683 | 0.09 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_+_117910047 | 0.09 |
ENST00000356554.3
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr10_+_124134201 | 0.09 |
ENST00000368990.3
ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr3_-_49459865 | 0.09 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr2_-_240969906 | 0.08 |
ENST00000402971.2
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2 |
| chr1_-_54411255 | 0.08 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr5_+_121647877 | 0.08 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr11_-_47546220 | 0.08 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr11_-_65626797 | 0.08 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr8_-_10588010 | 0.08 |
ENST00000304501.1
|
SOX7
|
SRY (sex determining region Y)-box 7 |
| chrX_+_154114635 | 0.08 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr16_-_3422283 | 0.08 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr9_+_104161123 | 0.08 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr3_-_58196939 | 0.08 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr8_+_120428546 | 0.08 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr10_+_64564469 | 0.08 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr12_-_14721283 | 0.08 |
ENST00000240617.5
|
PLBD1
|
phospholipase B domain containing 1 |
| chr21_+_44313375 | 0.08 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr13_+_37006421 | 0.08 |
ENST00000255465.4
|
CCNA1
|
cyclin A1 |
| chr20_-_47804894 | 0.08 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr5_-_131826457 | 0.08 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr2_-_47168906 | 0.08 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr11_+_60635035 | 0.08 |
ENST00000278853.5
|
ZP1
|
zona pellucida glycoprotein 1 (sperm receptor) |
| chr22_-_36925124 | 0.08 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr16_-_70713928 | 0.08 |
ENST00000576338.1
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_+_116519112 | 0.07 |
ENST00000369503.4
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr20_-_43589109 | 0.07 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr11_+_61717535 | 0.07 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr20_+_4129496 | 0.07 |
ENST00000346595.2
|
SMOX
|
spermine oxidase |
| chr7_-_75241096 | 0.07 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr8_-_12612962 | 0.07 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr3_-_113464906 | 0.07 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr5_+_72794233 | 0.07 |
ENST00000335895.8
ENST00000380591.3 ENST00000507081.2 |
BTF3
|
basic transcription factor 3 |
| chr6_+_36098363 | 0.07 |
ENST00000373759.1
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr12_+_12966250 | 0.07 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr4_-_186347099 | 0.07 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr11_-_77123065 | 0.07 |
ENST00000530617.1
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr20_-_33543546 | 0.07 |
ENST00000216951.2
|
GSS
|
glutathione synthetase |
| chr4_-_48908737 | 0.07 |
ENST00000381464.2
|
OCIAD2
|
OCIA domain containing 2 |
| chr18_+_34409069 | 0.07 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr12_+_69202795 | 0.07 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr5_+_121647386 | 0.07 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr18_-_34409116 | 0.07 |
ENST00000334295.4
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr1_-_155532484 | 0.07 |
ENST00000368346.3
ENST00000548830.1 |
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr1_+_114447763 | 0.07 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr10_+_96305610 | 0.07 |
ENST00000371332.4
ENST00000239026.6 |
HELLS
|
helicase, lymphoid-specific |
| chr11_+_100558384 | 0.06 |
ENST00000524892.2
ENST00000298815.8 |
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr6_+_76458990 | 0.06 |
ENST00000369977.3
|
MYO6
|
myosin VI |
| chr2_-_241759622 | 0.06 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr2_+_11295498 | 0.06 |
ENST00000295083.3
ENST00000441908.2 |
PQLC3
|
PQ loop repeat containing 3 |
| chr19_-_55690758 | 0.06 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr6_+_4776580 | 0.06 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chr8_-_23021533 | 0.06 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr6_-_138428613 | 0.06 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr12_+_99038998 | 0.06 |
ENST00000359972.2
ENST00000357310.1 ENST00000339433.3 ENST00000333991.1 |
APAF1
|
apoptotic peptidase activating factor 1 |
| chr9_-_140115775 | 0.06 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr1_-_153588334 | 0.06 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr14_+_50999744 | 0.06 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr9_+_80912059 | 0.06 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr3_-_49459878 | 0.06 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr10_+_70661014 | 0.06 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr13_-_20735178 | 0.06 |
ENST00000241125.3
|
GJA3
|
gap junction protein, alpha 3, 46kDa |
| chr12_-_6451014 | 0.06 |
ENST00000366159.4
ENST00000539372.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr6_+_52285046 | 0.06 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr20_+_57467204 | 0.06 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus |
| chr11_+_33279850 | 0.06 |
ENST00000531504.1
ENST00000456517.1 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr14_+_97263641 | 0.06 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr16_+_57406437 | 0.06 |
ENST00000564948.1
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr14_+_24702127 | 0.06 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr15_-_42386752 | 0.06 |
ENST00000290472.3
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic) |
| chr4_+_71768043 | 0.06 |
ENST00000502869.1
ENST00000309395.2 ENST00000396051.2 |
MOB1B
|
MOB kinase activator 1B |
| chr1_+_25599018 | 0.06 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr15_-_30685563 | 0.06 |
ENST00000401522.3
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr5_-_95297534 | 0.06 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr18_-_34408693 | 0.06 |
ENST00000587382.1
ENST00000589049.1 ENST00000587129.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr1_+_243419306 | 0.06 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr12_+_124086632 | 0.06 |
ENST00000238146.4
ENST00000538744.1 |
DDX55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr16_+_15744078 | 0.05 |
ENST00000396354.1
ENST00000570727.1 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr20_-_45035198 | 0.05 |
ENST00000372176.1
|
ELMO2
|
engulfment and cell motility 2 |
| chr19_+_489140 | 0.05 |
ENST00000587541.1
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr15_-_102285913 | 0.05 |
ENST00000558592.1
|
RP11-89K11.1
|
Uncharacterized protein |
| chr5_+_141348755 | 0.05 |
ENST00000506004.1
ENST00000507291.1 |
RNF14
|
ring finger protein 14 |
| chr1_+_42846443 | 0.05 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr11_-_59383617 | 0.05 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr11_+_125757556 | 0.05 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr1_+_54411715 | 0.05 |
ENST00000371370.3
ENST00000371368.1 |
LRRC42
|
leucine rich repeat containing 42 |
| chr1_+_54411995 | 0.05 |
ENST00000319223.4
ENST00000444987.1 |
LRRC42
|
leucine rich repeat containing 42 |
| chr11_-_3078838 | 0.05 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr19_-_5567996 | 0.05 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr1_-_213189168 | 0.05 |
ENST00000366962.3
ENST00000360506.2 |
ANGEL2
|
angel homolog 2 (Drosophila) |
| chr4_-_10117949 | 0.05 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr6_+_52285131 | 0.05 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr1_+_28206150 | 0.05 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chrX_-_70474377 | 0.05 |
ENST00000373978.1
ENST00000373981.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr22_-_36924944 | 0.05 |
ENST00000405442.1
ENST00000402116.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr2_-_37193606 | 0.05 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr10_+_126490354 | 0.05 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr1_+_151584544 | 0.05 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr19_+_18485509 | 0.05 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_110283138 | 0.05 |
ENST00000256594.3
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr6_+_24403144 | 0.05 |
ENST00000274747.7
ENST00000543597.1 ENST00000535061.1 ENST00000378353.1 ENST00000378386.3 ENST00000443868.2 |
MRS2
|
MRS2 magnesium transporter |
| chrX_+_134478706 | 0.05 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr1_-_54519134 | 0.05 |
ENST00000371341.1
|
TMEM59
|
transmembrane protein 59 |
| chr15_+_41913690 | 0.05 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr1_+_25598872 | 0.05 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP53
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.0 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |