Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
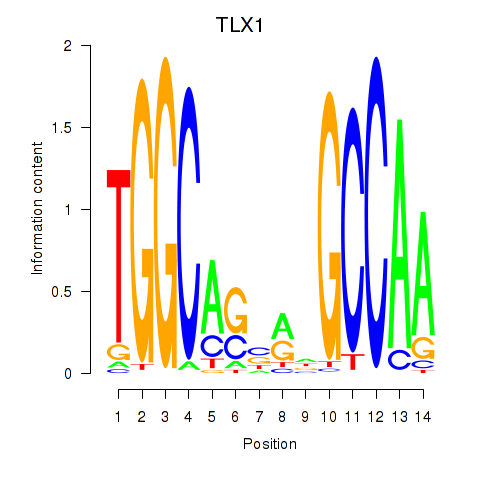
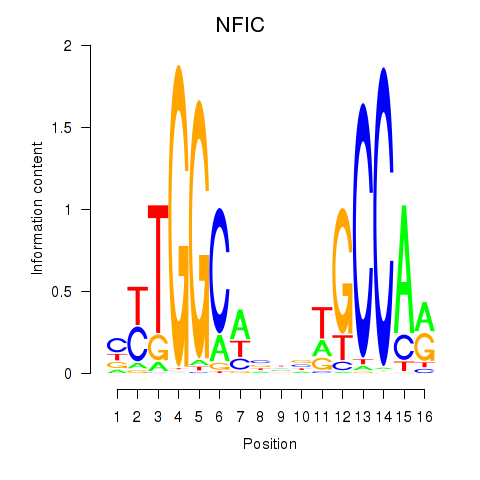
Results for TLX1_NFIC
Z-value: 1.15
Transcription factors associated with TLX1_NFIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX1
|
ENSG00000107807.8 | T cell leukemia homeobox 1 |
|
NFIC
|
ENSG00000141905.13 | nuclear factor I C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIC | hg19_v2_chr19_+_3366547_3366619 | 0.69 | 1.3e-01 | Click! |
| TLX1 | hg19_v2_chr10_+_102891048_102891078 | 0.45 | 3.7e-01 | Click! |
Activity profile of TLX1_NFIC motif
Sorted Z-values of TLX1_NFIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79086088 | 2.13 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr7_+_22766766 | 1.45 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chrX_-_40005865 | 0.91 |
ENST00000412952.1
|
BCOR
|
BCL6 corepressor |
| chr8_+_61822605 | 0.83 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr21_+_17792672 | 0.78 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_59010565 | 0.55 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr14_-_85996332 | 0.53 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr6_-_33663474 | 0.51 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr1_-_153363452 | 0.47 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr7_+_143078652 | 0.47 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr19_+_58111241 | 0.44 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr3_+_46742823 | 0.42 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr2_+_179317994 | 0.42 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr21_+_33784957 | 0.41 |
ENST00000401402.3
ENST00000382699.3 |
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr7_+_143079000 | 0.39 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr21_+_33784670 | 0.39 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr10_-_100174900 | 0.38 |
ENST00000370575.4
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr17_+_79369249 | 0.37 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr19_+_40871734 | 0.36 |
ENST00000359274.5
|
PLD3
|
phospholipase D family, member 3 |
| chr19_+_10397648 | 0.35 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr16_+_226658 | 0.35 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr6_+_132129151 | 0.34 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr8_-_42065187 | 0.34 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr9_-_98268883 | 0.33 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr13_-_40924439 | 0.33 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr3_-_131756559 | 0.32 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr16_+_30751953 | 0.32 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr12_-_109797249 | 0.32 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr19_+_10397621 | 0.31 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr8_-_42065075 | 0.31 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr20_+_48884002 | 0.31 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr1_-_154178803 | 0.31 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr15_+_89182178 | 0.31 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_+_30662085 | 0.31 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr6_+_31021973 | 0.31 |
ENST00000570223.1
ENST00000566475.1 ENST00000426185.1 |
HCG22
|
HLA complex group 22 |
| chr1_+_32674675 | 0.31 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr20_-_60942361 | 0.31 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr19_+_3708376 | 0.30 |
ENST00000539908.2
|
TJP3
|
tight junction protein 3 |
| chr19_-_42927251 | 0.30 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr17_-_41910505 | 0.30 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr12_-_109025849 | 0.29 |
ENST00000228463.6
|
SELPLG
|
selectin P ligand |
| chr19_+_3708338 | 0.29 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr1_-_200992827 | 0.29 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr14_-_50999373 | 0.29 |
ENST00000554273.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_-_145701718 | 0.28 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr15_+_89182156 | 0.28 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr9_+_102584128 | 0.28 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr1_+_110453203 | 0.28 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr10_+_106034637 | 0.28 |
ENST00000401888.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr16_+_75182376 | 0.28 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chrX_+_27826107 | 0.28 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr19_+_45312310 | 0.28 |
ENST00000589651.1
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr16_-_67450325 | 0.27 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chrX_-_54522558 | 0.27 |
ENST00000375135.3
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr17_-_74533734 | 0.27 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr10_+_102758105 | 0.26 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr1_+_153747746 | 0.26 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr11_-_66313699 | 0.26 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr4_-_110723335 | 0.26 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr11_-_65325664 | 0.25 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chrX_-_102319092 | 0.25 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr4_-_110723134 | 0.25 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr2_+_219187871 | 0.25 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr20_-_60942326 | 0.25 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr8_+_102064237 | 0.25 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1 |
| chr9_-_130617029 | 0.24 |
ENST00000373203.4
|
ENG
|
endoglin |
| chr2_+_7017796 | 0.24 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr16_-_2264779 | 0.24 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr16_-_69385681 | 0.24 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr3_-_50340996 | 0.24 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr6_-_53013620 | 0.24 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr15_+_75487984 | 0.24 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr8_-_145331153 | 0.23 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr17_+_1633755 | 0.23 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr18_-_53735601 | 0.23 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr22_+_50639408 | 0.23 |
ENST00000380903.2
|
SELO
|
Selenoprotein O |
| chrX_+_46937745 | 0.23 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr19_+_45312347 | 0.23 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr17_-_37764128 | 0.23 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr22_-_39640756 | 0.23 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr7_-_16844611 | 0.22 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr7_-_5463175 | 0.22 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chrX_-_53310791 | 0.22 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr8_+_104384616 | 0.22 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr1_+_27153173 | 0.22 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr5_-_1112141 | 0.22 |
ENST00000264930.5
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporter), member 7 |
| chr14_-_50999190 | 0.22 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr14_+_73704201 | 0.22 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr19_-_12946236 | 0.22 |
ENST00000589272.1
ENST00000393233.2 |
RTBDN
|
retbindin |
| chr6_-_16761678 | 0.22 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr19_+_40195101 | 0.21 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr9_+_34646651 | 0.21 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr18_-_74728998 | 0.21 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr17_+_7461613 | 0.21 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_+_135160844 | 0.21 |
ENST00000423766.1
ENST00000458230.1 |
PRAP1
|
proline-rich acidic protein 1 |
| chrX_+_49126294 | 0.21 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr22_-_31536480 | 0.21 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr1_+_110453514 | 0.20 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr3_+_48264816 | 0.19 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr21_+_46875395 | 0.19 |
ENST00000355480.5
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr3_+_52350335 | 0.19 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr11_-_111783919 | 0.19 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr17_-_7082668 | 0.19 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_+_38032216 | 0.19 |
ENST00000416303.1
|
VILL
|
villin-like |
| chr17_+_7462103 | 0.19 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_+_28996114 | 0.19 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr1_+_61547405 | 0.19 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr11_-_118789613 | 0.19 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr21_+_46875424 | 0.19 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr4_-_110723194 | 0.19 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr9_-_130616915 | 0.18 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr14_+_105939276 | 0.18 |
ENST00000483017.3
|
CRIP2
|
cysteine-rich protein 2 |
| chr7_+_150065278 | 0.18 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr4_-_84255935 | 0.18 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr16_+_30662184 | 0.17 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr3_-_53880401 | 0.17 |
ENST00000315251.6
|
CHDH
|
choline dehydrogenase |
| chr7_-_16840820 | 0.17 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr3_-_12200851 | 0.17 |
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr11_-_65325430 | 0.17 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr19_-_40324255 | 0.17 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr14_+_24867992 | 0.17 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr1_+_113217309 | 0.17 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr3_+_186915274 | 0.17 |
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1 |
| chr6_+_31950150 | 0.16 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr21_+_17443521 | 0.16 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_24772455 | 0.16 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr1_-_109399682 | 0.16 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr20_-_34025999 | 0.16 |
ENST00000374369.3
|
GDF5
|
growth differentiation factor 5 |
| chr7_+_143013198 | 0.16 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr1_-_32264250 | 0.16 |
ENST00000528579.1
|
SPOCD1
|
SPOC domain containing 1 |
| chr6_-_43595039 | 0.16 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr21_+_17791648 | 0.16 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr18_+_43304092 | 0.16 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr11_+_61717279 | 0.16 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr14_-_76447336 | 0.15 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr17_-_4806369 | 0.15 |
ENST00000293780.4
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon (muscle) |
| chr15_-_40213080 | 0.15 |
ENST00000561100.1
|
GPR176
|
G protein-coupled receptor 176 |
| chr11_+_61717336 | 0.15 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chrX_-_154688276 | 0.15 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr19_-_33716750 | 0.15 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr5_+_92919043 | 0.15 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr1_-_32264356 | 0.14 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr2_+_234296792 | 0.14 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr1_-_44497024 | 0.14 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_+_91446157 | 0.14 |
ENST00000559717.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr12_-_48744554 | 0.14 |
ENST00000544117.2
ENST00000548932.1 ENST00000549125.1 ENST00000301042.3 ENST00000547026.1 |
ZNF641
|
zinc finger protein 641 |
| chr19_+_1491144 | 0.14 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chrX_-_153744434 | 0.14 |
ENST00000369643.1
ENST00000393572.1 |
FAM3A
|
family with sequence similarity 3, member A |
| chr6_+_123100853 | 0.14 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr15_+_75105057 | 0.13 |
ENST00000309664.5
ENST00000562810.1 |
LMAN1L
|
lectin, mannose-binding, 1 like |
| chr2_-_217560248 | 0.13 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr16_-_19896220 | 0.13 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr12_-_54813229 | 0.13 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr17_+_78193443 | 0.13 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr2_+_74229812 | 0.13 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chrX_-_153744507 | 0.13 |
ENST00000442929.1
ENST00000426266.1 ENST00000359889.5 ENST00000369641.3 ENST00000447601.2 ENST00000434658.2 |
FAM3A
|
family with sequence similarity 3, member A |
| chr11_-_71752100 | 0.13 |
ENST00000542977.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr16_+_30662050 | 0.13 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr14_-_24047965 | 0.13 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr2_-_25475120 | 0.13 |
ENST00000380746.4
ENST00000402667.1 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr20_-_39317868 | 0.13 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr12_+_94656297 | 0.13 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr19_+_17579556 | 0.12 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr19_+_17326521 | 0.12 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr2_-_166930131 | 0.12 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr6_-_34524049 | 0.12 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chrX_+_48681768 | 0.12 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr8_+_22022800 | 0.12 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr1_+_152748848 | 0.12 |
ENST00000334371.2
|
LCE1F
|
late cornified envelope 1F |
| chr3_+_184080790 | 0.12 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_-_74533963 | 0.12 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr9_+_34646624 | 0.12 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr6_-_31550192 | 0.12 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr7_-_45128472 | 0.12 |
ENST00000490531.2
|
NACAD
|
NAC alpha domain containing |
| chr1_-_16539094 | 0.12 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr17_-_73874654 | 0.12 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr11_-_57089671 | 0.11 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr6_+_31949801 | 0.11 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr15_-_40633101 | 0.11 |
ENST00000559313.1
|
C15orf52
|
chromosome 15 open reading frame 52 |
| chr7_-_1595107 | 0.11 |
ENST00000414730.1
|
TMEM184A
|
transmembrane protein 184A |
| chr11_-_111784005 | 0.11 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_9903938 | 0.11 |
ENST00000511616.1
|
CTD-2143L24.1
|
CTD-2143L24.1 |
| chr1_+_110453608 | 0.11 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_-_67771513 | 0.11 |
ENST00000227471.2
|
UNC93B1
|
unc-93 homolog B1 (C. elegans) |
| chr7_+_73242069 | 0.11 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr7_+_18330035 | 0.11 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chrX_-_152760934 | 0.11 |
ENST00000370210.1
ENST00000421080.2 |
HAUS7
|
HAUS augmin-like complex, subunit 7 |
| chr19_-_44259136 | 0.11 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr8_-_141728760 | 0.11 |
ENST00000430260.2
|
PTK2
|
protein tyrosine kinase 2 |
| chr20_+_9049742 | 0.11 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chrX_+_154114635 | 0.11 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr11_-_64764435 | 0.11 |
ENST00000534177.1
ENST00000301887.4 |
BATF2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr22_+_46449674 | 0.11 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr1_-_32169920 | 0.11 |
ENST00000373672.3
ENST00000373668.3 |
COL16A1
|
collagen, type XVI, alpha 1 |
| chr5_+_150020240 | 0.10 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr15_-_49255632 | 0.10 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr1_+_113217043 | 0.10 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_+_113217345 | 0.10 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_+_7461580 | 0.10 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_+_6644443 | 0.10 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX1_NFIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.9 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.6 | GO:1904124 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.6 | GO:0060481 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of sperm motility(GO:1901318) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.4 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.3 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.5 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 0.2 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.4 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.3 | GO:1900164 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.3 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.6 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 0.6 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.5 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.1 | 1.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.3 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |