Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
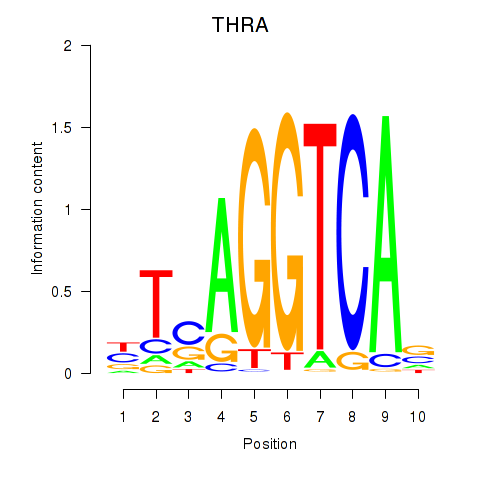
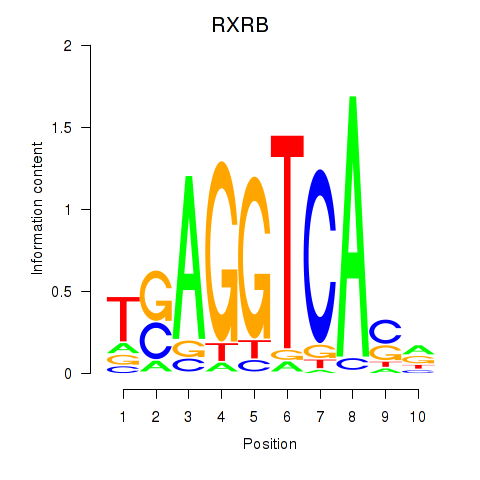
Results for THRA_RXRB
Z-value: 0.23
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RXRB | hg19_v2_chr6_-_33168391_33168465 | -0.28 | 6.0e-01 | Click! |
| THRA | hg19_v2_chr17_+_38219063_38219154 | -0.06 | 9.2e-01 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_162887054 | 0.21 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr1_-_12679171 | 0.20 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr22_+_23264766 | 0.18 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr5_+_149865377 | 0.18 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_+_86999516 | 0.17 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_+_229440129 | 0.13 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr10_+_77056181 | 0.12 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr6_+_13272904 | 0.10 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_-_33402506 | 0.10 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr10_+_26986582 | 0.09 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr20_+_33146510 | 0.09 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr5_-_162887071 | 0.09 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr12_-_124873357 | 0.09 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr1_-_11907829 | 0.07 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr12_+_25205155 | 0.07 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr9_-_88874519 | 0.07 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr3_-_49726104 | 0.07 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr22_+_29138013 | 0.07 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr18_-_19748379 | 0.06 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr12_+_56477093 | 0.06 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_1553346 | 0.06 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr6_+_35773070 | 0.06 |
ENST00000373853.1
ENST00000360215.1 |
LHFPL5
|
lipoma HMGIC fusion partner-like 5 |
| chr19_-_58485895 | 0.06 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr22_-_33968239 | 0.06 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr19_+_32836499 | 0.06 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr17_+_38121772 | 0.06 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr15_-_75017711 | 0.06 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr2_-_55459485 | 0.05 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr19_+_49467232 | 0.05 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chrX_-_101397433 | 0.05 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chr15_-_93965805 | 0.05 |
ENST00000556708.1
|
RP11-164C12.2
|
RP11-164C12.2 |
| chr12_-_111926342 | 0.05 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr19_-_5838734 | 0.05 |
ENST00000532464.1
ENST00000528505.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr12_+_41221975 | 0.05 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr19_+_54371114 | 0.05 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr16_-_11367452 | 0.05 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr3_+_14989186 | 0.05 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_203274418 | 0.05 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr19_-_4517613 | 0.04 |
ENST00000301286.3
|
PLIN4
|
perilipin 4 |
| chr15_-_34629922 | 0.04 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr15_+_45422131 | 0.04 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr15_-_34630234 | 0.04 |
ENST00000558667.1
ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr17_-_1928621 | 0.04 |
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1 |
| chr12_+_7072354 | 0.04 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr3_-_100566492 | 0.04 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr8_+_97597148 | 0.04 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr19_-_44809121 | 0.04 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr4_+_20255123 | 0.04 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chrX_+_102840408 | 0.04 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr6_-_34664612 | 0.04 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr10_+_99332198 | 0.04 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr15_-_45422056 | 0.04 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr11_-_59578202 | 0.04 |
ENST00000300151.4
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr19_-_52227221 | 0.04 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr1_+_41204506 | 0.04 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr22_+_30821732 | 0.04 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr14_+_24616588 | 0.04 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr6_-_31782813 | 0.04 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr16_+_85646763 | 0.03 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr19_+_39989580 | 0.03 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr12_+_120875910 | 0.03 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr1_+_26517052 | 0.03 |
ENST00000338855.2
ENST00000456354.2 |
CATSPER4
|
cation channel, sperm associated 4 |
| chr20_-_4055812 | 0.03 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr3_+_45067659 | 0.03 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr21_+_17566643 | 0.03 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_30723003 | 0.03 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr7_+_128431444 | 0.03 |
ENST00000459946.1
ENST00000378685.4 ENST00000464832.1 ENST00000472049.1 ENST00000488925.1 |
CCDC136
|
coiled-coil domain containing 136 |
| chr11_-_119599794 | 0.03 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr6_+_30295036 | 0.03 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr15_-_41408409 | 0.03 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chrX_+_48681768 | 0.03 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr9_+_2017063 | 0.03 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_61389168 | 0.03 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr7_+_102389434 | 0.03 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr17_-_74489215 | 0.03 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr15_+_45422178 | 0.03 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr17_-_41132088 | 0.03 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr7_-_73153161 | 0.03 |
ENST00000395147.4
|
ABHD11
|
abhydrolase domain containing 11 |
| chr17_+_4981535 | 0.03 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chrX_+_70521584 | 0.03 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr1_-_151431674 | 0.03 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr7_-_56160666 | 0.03 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr2_+_97454321 | 0.03 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr3_+_138153451 | 0.03 |
ENST00000389567.4
ENST00000289135.4 |
ESYT3
|
extended synaptotagmin-like protein 3 |
| chr17_-_7082668 | 0.03 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr19_-_49565254 | 0.03 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chrX_-_152989798 | 0.02 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr5_+_139927213 | 0.02 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr16_-_70729496 | 0.02 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr10_+_95372289 | 0.02 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr9_+_35042205 | 0.02 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr1_-_173886491 | 0.02 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr11_-_795286 | 0.02 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr9_+_140145713 | 0.02 |
ENST00000388931.3
ENST00000412566.1 |
C9orf173
|
chromosome 9 open reading frame 173 |
| chr1_-_205719295 | 0.02 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr4_+_26165074 | 0.02 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr22_-_20255212 | 0.02 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr19_+_6739662 | 0.02 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr11_+_1856034 | 0.02 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr17_+_44928946 | 0.02 |
ENST00000290015.2
ENST00000393461.2 |
WNT9B
|
wingless-type MMTV integration site family, member 9B |
| chr20_+_19870167 | 0.02 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr5_-_131563474 | 0.02 |
ENST00000417528.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr22_+_17082732 | 0.02 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr19_-_41196458 | 0.02 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr10_+_99332529 | 0.02 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr13_-_46756351 | 0.02 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr11_+_33563618 | 0.02 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr22_+_23487513 | 0.02 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr17_-_4843206 | 0.02 |
ENST00000576951.1
|
SLC25A11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr6_+_43457317 | 0.02 |
ENST00000438588.2
|
TJAP1
|
tight junction associated protein 1 (peripheral) |
| chr10_-_105218645 | 0.02 |
ENST00000329905.5
|
CALHM1
|
calcium homeostasis modulator 1 |
| chr4_+_169842707 | 0.02 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_149829244 | 0.02 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr2_+_45168875 | 0.02 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr10_+_63808970 | 0.02 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr13_+_113030658 | 0.02 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr14_-_103523745 | 0.02 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr17_-_80231557 | 0.02 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr16_-_67700594 | 0.02 |
ENST00000602644.1
ENST00000243878.4 |
ENKD1
|
enkurin domain containing 1 |
| chr3_-_195270162 | 0.02 |
ENST00000438848.1
ENST00000328432.3 |
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr22_-_42486747 | 0.02 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr2_+_113670548 | 0.02 |
ENST00000263326.3
ENST00000352179.3 ENST00000349806.3 ENST00000353225.3 |
IL37
|
interleukin 37 |
| chr1_-_91487806 | 0.02 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr8_-_53322303 | 0.02 |
ENST00000276480.7
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr4_+_126237554 | 0.02 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr22_-_20138302 | 0.02 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr1_-_151119087 | 0.02 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr1_+_179851893 | 0.02 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chrX_+_101380642 | 0.02 |
ENST00000372780.1
ENST00000329035.2 |
TCEAL2
|
transcription elongation factor A (SII)-like 2 |
| chr20_+_1875110 | 0.02 |
ENST00000400068.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr2_+_210518057 | 0.02 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr16_+_2106134 | 0.02 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr1_-_204121298 | 0.02 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_151431647 | 0.02 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr3_-_49726486 | 0.02 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr19_+_16771936 | 0.02 |
ENST00000187762.2
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chrX_+_102862834 | 0.02 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr5_-_131563501 | 0.02 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr1_-_109940550 | 0.02 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr2_-_192016276 | 0.02 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr11_-_117699413 | 0.02 |
ENST00000528014.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr2_+_39117010 | 0.02 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr5_-_149829294 | 0.02 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr12_+_95611569 | 0.02 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_41196534 | 0.02 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr12_-_48744554 | 0.02 |
ENST00000544117.2
ENST00000548932.1 ENST00000549125.1 ENST00000301042.3 ENST00000547026.1 |
ZNF641
|
zinc finger protein 641 |
| chr3_-_38992052 | 0.02 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr11_+_1860200 | 0.02 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr15_-_41408339 | 0.02 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr1_-_144994909 | 0.02 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_+_104178946 | 0.02 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr8_+_98881268 | 0.02 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr8_+_37553261 | 0.02 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chrX_-_39956656 | 0.02 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr2_+_231860830 | 0.02 |
ENST00000424440.1
ENST00000452881.1 ENST00000433428.2 ENST00000455816.1 ENST00000440792.1 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr4_+_26322409 | 0.02 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_141105235 | 0.02 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_230787201 | 0.02 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr1_-_38230738 | 0.02 |
ENST00000427468.2
ENST00000373048.4 ENST00000319637.6 |
EPHA10
|
EPH receptor A10 |
| chr17_+_38337491 | 0.02 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_+_70876891 | 0.01 |
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr16_-_88752889 | 0.01 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr3_+_172361483 | 0.01 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr1_+_236558694 | 0.01 |
ENST00000359362.5
|
EDARADD
|
EDAR-associated death domain |
| chr17_-_36904437 | 0.01 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr19_+_49109990 | 0.01 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr1_-_149900122 | 0.01 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr14_+_67999999 | 0.01 |
ENST00000329153.5
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr9_+_75263565 | 0.01 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr12_-_11139511 | 0.01 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr14_+_74551650 | 0.01 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr2_+_220491973 | 0.01 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr11_+_7618413 | 0.01 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_57914742 | 0.01 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr17_-_27503770 | 0.01 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr10_+_81065975 | 0.01 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr17_+_42264322 | 0.01 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr5_-_131562935 | 0.01 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr18_+_19668021 | 0.01 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr19_+_1248547 | 0.01 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr19_+_58898627 | 0.01 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr12_-_110011288 | 0.01 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr7_+_26591441 | 0.01 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr6_+_30614779 | 0.01 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr1_-_204121102 | 0.01 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr20_+_33464238 | 0.01 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_+_41249539 | 0.01 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chrX_+_23928500 | 0.01 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr11_-_795400 | 0.01 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr20_+_1875378 | 0.01 |
ENST00000356025.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr11_-_119066545 | 0.01 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr13_-_41635512 | 0.01 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr12_+_120875887 | 0.01 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr11_+_65190245 | 0.01 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr7_-_73153122 | 0.01 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr16_+_28875126 | 0.01 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr20_+_42193856 | 0.01 |
ENST00000412111.1
ENST00000423407.3 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr14_+_32964258 | 0.01 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_-_42698433 | 0.01 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr16_+_69796209 | 0.01 |
ENST00000359154.2
ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.0 | GO:1902623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |