|
chr2_+_228678550
Show fit
|
1.34 |
ENST00000409189.3
ENST00000358813.4
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr5_-_150466692
Show fit
|
0.60 |
ENST00000315050.7
ENST00000523338.1
ENST00000522100.1
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr16_+_30194916
Show fit
|
0.51 |
ENST00000570045.1
ENST00000565497.1
ENST00000570244.1
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr11_+_18287721
Show fit
|
0.48 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1
|
|
chr11_+_18287801
Show fit
|
0.47 |
ENST00000532858.1
ENST00000405158.2
|
SAA1
|
serum amyloid A1
|
|
chr18_+_54318893
Show fit
|
0.46 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7
|
|
chr8_-_42065075
Show fit
|
0.45 |
ENST00000429089.2
ENST00000519510.1
ENST00000429710.2
ENST00000524009.1
|
PLAT
|
plasminogen activator, tissue
|
|
chr1_+_79086088
Show fit
|
0.39 |
ENST00000370751.5
ENST00000342282.3
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr8_+_72755367
Show fit
|
0.37 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548
|
|
chrX_+_49019061
Show fit
|
0.36 |
ENST00000376339.1
ENST00000425661.2
ENST00000458388.1
ENST00000412696.2
|
MAGIX
|
MAGI family member, X-linked
|
|
chr7_+_76101379
Show fit
|
0.36 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr5_-_66942617
Show fit
|
0.33 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5
|
|
chr1_+_35734616
Show fit
|
0.33 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr2_-_36779411
Show fit
|
0.33 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein
|
|
chr6_+_138188551
Show fit
|
0.32 |
ENST00000237289.4
ENST00000433680.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_113594279
Show fit
|
0.32 |
ENST00000416750.1
ENST00000418817.1
ENST00000263341.2
|
IL1B
|
interleukin 1, beta
|
|
chr11_-_26743546
Show fit
|
0.32 |
ENST00000280467.6
ENST00000396005.3
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12
|
|
chr1_-_11118896
Show fit
|
0.30 |
ENST00000465788.1
|
SRM
|
spermidine synthase
|
|
chr6_-_72129806
Show fit
|
0.30 |
ENST00000413945.1
ENST00000602878.1
ENST00000436803.1
ENST00000421704.1
ENST00000441570.1
|
LINC00472
|
long intergenic non-protein coding RNA 472
|
|
chr17_-_39661947
Show fit
|
0.29 |
ENST00000590425.1
|
KRT13
|
keratin 13
|
|
chr12_-_62653903
Show fit
|
0.29 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr5_-_150467221
Show fit
|
0.29 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr5_-_74348371
Show fit
|
0.28 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2
|
|
chr11_-_128457446
Show fit
|
0.27 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1
|
|
chr14_-_24732403
Show fit
|
0.27 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1
|
|
chr19_-_51538148
Show fit
|
0.25 |
ENST00000319590.4
ENST00000250351.4
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr14_-_24732368
Show fit
|
0.25 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1
|
|
chr4_+_8183793
Show fit
|
0.25 |
ENST00000509119.1
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr13_+_76413852
Show fit
|
0.24 |
ENST00000533809.2
|
LMO7
|
LIM domain 7
|
|
chr14_-_75083313
Show fit
|
0.24 |
ENST00000556652.1
ENST00000555313.1
|
CTD-2207P18.2
|
CTD-2207P18.2
|
|
chr12_+_25348139
Show fit
|
0.23 |
ENST00000557540.2
ENST00000381356.4
|
LYRM5
|
LYR motif containing 5
|
|
chr5_-_115890554
Show fit
|
0.23 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr4_+_88896819
Show fit
|
0.23 |
ENST00000237623.7
ENST00000395080.3
ENST00000508233.1
ENST00000360804.4
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_+_49621658
Show fit
|
0.23 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr3_+_122920847
Show fit
|
0.22 |
ENST00000466519.1
ENST00000480631.1
ENST00000491366.1
ENST00000487572.1
|
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae)
|
|
chr9_-_35812140
Show fit
|
0.22 |
ENST00000497810.1
ENST00000396638.2
ENST00000484764.1
|
SPAG8
|
sperm associated antigen 8
|
|
chr3_-_127541194
Show fit
|
0.21 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase
|
|
chr12_+_65672702
Show fit
|
0.21 |
ENST00000538045.1
ENST00000535239.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr19_-_51537982
Show fit
|
0.21 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr17_-_71223839
Show fit
|
0.21 |
ENST00000579872.1
ENST00000580032.1
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr15_-_43559055
Show fit
|
0.20 |
ENST00000220420.5
ENST00000349114.4
|
TGM5
|
transglutaminase 5
|
|
chr14_+_70078303
Show fit
|
0.20 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247
|
|
chr4_-_38807199
Show fit
|
0.20 |
ENST00000508364.1
|
TLR1
|
toll-like receptor 1
|
|
chr12_+_25348186
Show fit
|
0.20 |
ENST00000555711.1
ENST00000556885.1
ENST00000554266.1
ENST00000556351.1
ENST00000556927.1
ENST00000556402.1
ENST00000553788.1
|
LYRM5
|
LYR motif containing 5
|
|
chr8_-_82395461
Show fit
|
0.19 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr11_+_12132117
Show fit
|
0.19 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2
|
|
chr5_-_65018834
Show fit
|
0.19 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr3_+_23244579
Show fit
|
0.18 |
ENST00000452894.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr17_+_26369865
Show fit
|
0.18 |
ENST00000582037.1
|
NLK
|
nemo-like kinase
|
|
chr19_-_51538118
Show fit
|
0.18 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr8_+_71485681
Show fit
|
0.18 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1
|
|
chr3_-_127541679
Show fit
|
0.18 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase
|
|
chr14_+_21492331
Show fit
|
0.18 |
ENST00000533984.1
ENST00000532213.2
|
AL161668.5
|
AL161668.5
|
|
chr17_-_66287310
Show fit
|
0.18 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6
|
|
chr10_-_11574274
Show fit
|
0.18 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like
|
|
chr5_+_76012009
Show fit
|
0.18 |
ENST00000505600.1
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr19_-_51531210
Show fit
|
0.18 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr1_-_53163992
Show fit
|
0.18 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7
|
|
chr12_+_78224667
Show fit
|
0.18 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3
|
|
chr2_-_74779744
Show fit
|
0.18 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr20_-_31124186
Show fit
|
0.17 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr5_+_139055021
Show fit
|
0.17 |
ENST00000502716.1
ENST00000503511.1
|
CXXC5
|
CXXC finger protein 5
|
|
chr6_-_3456267
Show fit
|
0.17 |
ENST00000485307.1
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr6_-_3227877
Show fit
|
0.17 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chr14_+_100842735
Show fit
|
0.17 |
ENST00000554998.1
ENST00000402312.3
ENST00000335290.6
ENST00000554175.1
|
WDR25
|
WD repeat domain 25
|
|
chr16_-_30582888
Show fit
|
0.17 |
ENST00000563707.1
ENST00000567855.1
|
ZNF688
|
zinc finger protein 688
|
|
chr13_+_78109804
Show fit
|
0.17 |
ENST00000535157.1
|
SCEL
|
sciellin
|
|
chr15_+_75498355
Show fit
|
0.17 |
ENST00000567617.1
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr14_-_21493123
Show fit
|
0.17 |
ENST00000556147.1
ENST00000554489.1
ENST00000555657.1
ENST00000557274.1
ENST00000555158.1
ENST00000554833.1
ENST00000555384.1
ENST00000556420.1
ENST00000554893.1
ENST00000553503.1
ENST00000555733.1
ENST00000553867.1
ENST00000397856.3
ENST00000397855.3
ENST00000556008.1
ENST00000557182.1
ENST00000554483.1
ENST00000556688.1
ENST00000397853.3
ENST00000556329.2
ENST00000554143.1
ENST00000397851.2
ENST00000555142.1
ENST00000557676.1
ENST00000556924.1
|
NDRG2
|
NDRG family member 2
|
|
chr19_-_13044494
Show fit
|
0.16 |
ENST00000593021.1
ENST00000587981.1
ENST00000423140.2
ENST00000314606.4
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr15_+_44119159
Show fit
|
0.16 |
ENST00000263795.6
ENST00000381246.2
ENST00000452115.1
|
WDR76
|
WD repeat domain 76
|
|
chr21_+_44394620
Show fit
|
0.16 |
ENST00000291547.5
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr18_+_32556892
Show fit
|
0.16 |
ENST00000591734.1
ENST00000413393.1
ENST00000589180.1
ENST00000587359.1
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr12_-_71512027
Show fit
|
0.16 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein
|
|
chr11_+_89867803
Show fit
|
0.15 |
ENST00000321955.4
ENST00000525171.1
ENST00000375944.3
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2
|
|
chr18_+_66465475
Show fit
|
0.15 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr6_-_10415218
Show fit
|
0.15 |
ENST00000466073.1
ENST00000498450.1
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_+_27505260
Show fit
|
0.15 |
ENST00000380075.2
ENST00000296098.4
|
TRIM54
|
tripartite motif containing 54
|
|
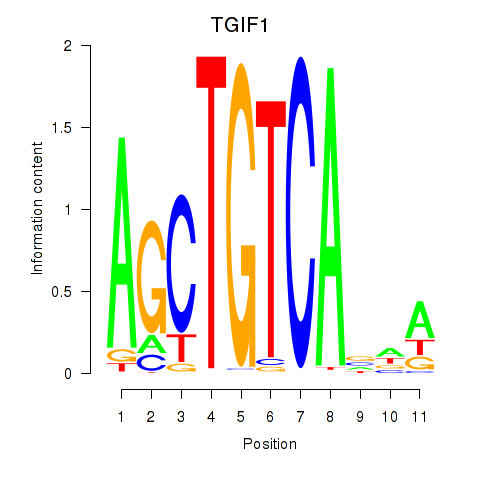
chr18_+_3450161
Show fit
|
0.15 |
ENST00000551402.1
ENST00000577543.1
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr3_-_127542021
Show fit
|
0.15 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase
|
|
chr1_+_36621697
Show fit
|
0.15 |
ENST00000373150.4
ENST00000373151.2
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr2_+_118846008
Show fit
|
0.15 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2
|
|
chr10_+_69869237
Show fit
|
0.15 |
ENST00000373675.3
|
MYPN
|
myopalladin
|
|
chr5_+_141348721
Show fit
|
0.15 |
ENST00000507163.1
ENST00000394519.1
|
RNF14
|
ring finger protein 14
|
|
chr2_-_73460334
Show fit
|
0.14 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1
|
|
chr13_+_78109884
Show fit
|
0.14 |
ENST00000377246.3
ENST00000349847.3
|
SCEL
|
sciellin
|
|
chr18_+_66465302
Show fit
|
0.14 |
ENST00000360242.5
ENST00000358653.5
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr3_-_127542051
Show fit
|
0.14 |
ENST00000398104.1
|
MGLL
|
monoglyceride lipase
|
|
chr1_+_152943122
Show fit
|
0.14 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4
|
|
chr19_-_10946871
Show fit
|
0.14 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1
|
|
chr7_+_101928416
Show fit
|
0.14 |
ENST00000444095.1
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr12_+_55248289
Show fit
|
0.14 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1
|
|
chr17_-_55911970
Show fit
|
0.14 |
ENST00000581805.1
ENST00000580960.1
|
RP11-60A24.3
|
RP11-60A24.3
|
|
chr4_+_55095428
Show fit
|
0.14 |
ENST00000508170.1
ENST00000512143.1
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr9_+_125133467
Show fit
|
0.14 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr2_-_179914760
Show fit
|
0.14 |
ENST00000420890.2
ENST00000409284.1
ENST00000443758.1
ENST00000446116.1
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr1_-_186649543
Show fit
|
0.14 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr17_-_46608272
Show fit
|
0.14 |
ENST00000577092.1
ENST00000239174.6
|
HOXB1
|
homeobox B1
|
|
chr18_+_54318616
Show fit
|
0.13 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7
|
|
chr6_+_88106840
Show fit
|
0.13 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164
|
|
chr20_+_35202909
Show fit
|
0.13 |
ENST00000558530.1
ENST00000558028.1
ENST00000560025.1
|
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough
TGFB-induced factor homeobox 2
|
|
chr3_+_169629354
Show fit
|
0.13 |
ENST00000428432.2
ENST00000335556.3
|
SAMD7
|
sterile alpha motif domain containing 7
|
|
chr12_-_76461249
Show fit
|
0.13 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr5_+_133984462
Show fit
|
0.13 |
ENST00000398844.2
ENST00000322887.4
|
SEC24A
|
SEC24 family member A
|
|
chr10_+_94608245
Show fit
|
0.13 |
ENST00000443748.2
ENST00000260762.6
|
EXOC6
|
exocyst complex component 6
|
|
chr8_+_145438870
Show fit
|
0.13 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B
|
|
chr6_+_30951487
Show fit
|
0.13 |
ENST00000486149.2
ENST00000376296.3
|
MUC21
|
mucin 21, cell surface associated
|
|
chr17_-_39661849
Show fit
|
0.13 |
ENST00000246635.3
ENST00000336861.3
ENST00000587544.1
ENST00000587435.1
|
KRT13
|
keratin 13
|
|
chr22_-_50219548
Show fit
|
0.13 |
ENST00000404034.1
|
BRD1
|
bromodomain containing 1
|
|
chr3_+_106959552
Show fit
|
0.13 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883
|
|
chr7_-_93520191
Show fit
|
0.13 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr19_+_7953417
Show fit
|
0.13 |
ENST00000600345.1
ENST00000598224.1
|
LRRC8E
|
leucine rich repeat containing 8 family, member E
|
|
chr1_-_193075180
Show fit
|
0.13 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2
|
|
chr8_+_38243821
Show fit
|
0.12 |
ENST00000519476.2
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr9_-_130635741
Show fit
|
0.12 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1
|
|
chr11_-_64052111
Show fit
|
0.12 |
ENST00000394532.3
ENST00000394531.3
ENST00000309032.3
|
BAD
|
BCL2-associated agonist of cell death
|
|
chrX_-_128788914
Show fit
|
0.12 |
ENST00000429967.1
ENST00000307484.6
|
APLN
|
apelin
|
|
chr18_+_54318566
Show fit
|
0.12 |
ENST00000589935.1
ENST00000357574.3
|
WDR7
|
WD repeat domain 7
|
|
chr16_-_30134524
Show fit
|
0.12 |
ENST00000395202.1
ENST00000395199.3
ENST00000263025.4
ENST00000322266.5
ENST00000403394.1
|
MAPK3
|
mitogen-activated protein kinase 3
|
|
chr4_-_105416039
Show fit
|
0.12 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4
|
|
chr5_-_159797627
Show fit
|
0.12 |
ENST00000393975.3
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chr2_-_18770802
Show fit
|
0.12 |
ENST00000416783.1
|
NT5C1B
|
5'-nucleotidase, cytosolic IB
|
|
chr11_+_842808
Show fit
|
0.12 |
ENST00000397397.2
ENST00000397411.2
ENST00000397396.1
|
TSPAN4
|
tetraspanin 4
|
|
chr12_+_65672423
Show fit
|
0.12 |
ENST00000355192.3
ENST00000308259.5
ENST00000540804.1
ENST00000535664.1
ENST00000541189.1
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr6_+_24126350
Show fit
|
0.12 |
ENST00000378491.4
ENST00000378478.1
ENST00000378477.2
|
NRSN1
|
neurensin 1
|
|
chr15_+_36887069
Show fit
|
0.12 |
ENST00000566807.1
ENST00000567389.1
ENST00000562877.1
|
C15orf41
|
chromosome 15 open reading frame 41
|
|
chr11_-_118023594
Show fit
|
0.11 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit
|
|
chr19_-_38085633
Show fit
|
0.11 |
ENST00000593133.1
ENST00000590751.1
ENST00000358744.3
ENST00000328550.2
ENST00000451802.2
|
ZNF571
|
zinc finger protein 571
|
|
chr2_-_74780176
Show fit
|
0.11 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr1_-_39407467
Show fit
|
0.11 |
ENST00000540558.1
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr10_-_99393242
Show fit
|
0.11 |
ENST00000370635.3
ENST00000478953.1
ENST00000335628.3
|
MORN4
|
MORN repeat containing 4
|
|
chr22_+_41347363
Show fit
|
0.11 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase
|
|
chr9_+_130911723
Show fit
|
0.11 |
ENST00000277480.2
ENST00000373013.2
ENST00000540948.1
|
LCN2
|
lipocalin 2
|
|
chr22_+_37959647
Show fit
|
0.11 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr17_+_78518617
Show fit
|
0.11 |
ENST00000537330.1
ENST00000570891.1
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr11_-_10822029
Show fit
|
0.11 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr18_-_67872891
Show fit
|
0.11 |
ENST00000454359.1
ENST00000437017.1
|
RTTN
|
rotatin
|
|
chr14_-_38028689
Show fit
|
0.11 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2
|
|
chr10_+_60028818
Show fit
|
0.11 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
chr16_-_57831676
Show fit
|
0.11 |
ENST00000465878.2
ENST00000539578.1
ENST00000561524.1
|
KIFC3
|
kinesin family member C3
|
|
chr3_+_113667354
Show fit
|
0.11 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr1_-_204329013
Show fit
|
0.11 |
ENST00000272203.3
ENST00000414478.1
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr1_+_153388993
Show fit
|
0.11 |
ENST00000368729.4
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chrX_+_153775869
Show fit
|
0.11 |
ENST00000424839.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr15_+_75498739
Show fit
|
0.11 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr16_+_2880254
Show fit
|
0.10 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B
|
|
chr3_+_122920767
Show fit
|
0.10 |
ENST00000492595.1
ENST00000473494.2
ENST00000481965.2
|
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae)
|
|
chr7_+_33945106
Show fit
|
0.10 |
ENST00000426693.1
|
BMPER
|
BMP binding endothelial regulator
|
|
chr4_+_24661479
Show fit
|
0.10 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2
|
|
chr11_-_8795787
Show fit
|
0.10 |
ENST00000528196.1
ENST00000533681.1
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_-_39407450
Show fit
|
0.10 |
ENST00000372990.1
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr21_+_43934633
Show fit
|
0.10 |
ENST00000398343.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1
|
|
chr1_+_74701062
Show fit
|
0.10 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr1_-_39395165
Show fit
|
0.10 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr15_-_82338460
Show fit
|
0.10 |
ENST00000558133.1
ENST00000329713.4
|
MEX3B
|
mex-3 RNA binding family member B
|
|
chr19_-_49865639
Show fit
|
0.10 |
ENST00000593945.1
ENST00000601519.1
ENST00000539846.1
ENST00000596757.1
ENST00000311227.2
|
TEAD2
|
TEA domain family member 2
|
|
chr1_+_205473720
Show fit
|
0.10 |
ENST00000429964.2
ENST00000506784.1
ENST00000360066.2
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr7_+_150811705
Show fit
|
0.10 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr11_+_842928
Show fit
|
0.10 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4
|
|
chr6_-_27840099
Show fit
|
0.10 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i
|
|
chr19_-_4182530
Show fit
|
0.10 |
ENST00000601571.1
ENST00000601488.1
ENST00000305232.6
ENST00000381935.3
ENST00000337491.2
|
SIRT6
|
sirtuin 6
|
|
chr1_+_26759295
Show fit
|
0.10 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr16_+_2880369
Show fit
|
0.10 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B
|
|
chr9_+_130911770
Show fit
|
0.10 |
ENST00000372998.1
|
LCN2
|
lipocalin 2
|
|
chr8_-_20161466
Show fit
|
0.10 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr22_-_37640277
Show fit
|
0.09 |
ENST00000401529.3
ENST00000249071.6
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr6_-_13487825
Show fit
|
0.09 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr8_+_102064237
Show fit
|
0.09 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1
|
|
chr20_+_61299155
Show fit
|
0.09 |
ENST00000451793.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chrX_-_71525742
Show fit
|
0.09 |
ENST00000450875.1
ENST00000417400.1
ENST00000431381.1
ENST00000445983.1
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr18_-_54318353
Show fit
|
0.09 |
ENST00000590954.1
ENST00000540155.1
|
TXNL1
|
thioredoxin-like 1
|
|
chr3_-_195538728
Show fit
|
0.09 |
ENST00000349607.4
ENST00000346145.4
|
MUC4
|
mucin 4, cell surface associated
|
|
chr21_+_43933946
Show fit
|
0.09 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1
|
|
chr2_+_173724771
Show fit
|
0.09 |
ENST00000538974.1
ENST00000540783.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr1_+_198608146
Show fit
|
0.09 |
ENST00000367376.2
ENST00000352140.3
ENST00000594404.1
ENST00000598951.1
ENST00000530727.1
ENST00000442510.2
ENST00000367367.4
ENST00000348564.6
ENST00000367364.1
ENST00000413409.2
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_48620147
Show fit
|
0.09 |
ENST00000303227.6
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr16_+_50775948
Show fit
|
0.09 |
ENST00000569681.1
ENST00000569418.1
ENST00000540145.1
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr6_+_32146268
Show fit
|
0.09 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase
|
|
chr3_-_33686925
Show fit
|
0.09 |
ENST00000485378.2
ENST00000313350.6
ENST00000487200.1
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr19_-_10946949
Show fit
|
0.09 |
ENST00000214869.2
ENST00000591695.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1
|
|
chr9_+_131873591
Show fit
|
0.08 |
ENST00000393370.2
ENST00000337738.1
ENST00000348141.5
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr5_+_141348640
Show fit
|
0.08 |
ENST00000540015.1
ENST00000506938.1
ENST00000394514.2
ENST00000512565.1
ENST00000394515.3
|
RNF14
|
ring finger protein 14
|
|
chr10_-_49812997
Show fit
|
0.08 |
ENST00000417912.2
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr19_+_49375649
Show fit
|
0.08 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr1_+_245318279
Show fit
|
0.08 |
ENST00000407071.2
|
KIF26B
|
kinesin family member 26B
|
|
chr16_+_30936971
Show fit
|
0.08 |
ENST00000565690.1
|
FBXL19
|
F-box and leucine-rich repeat protein 19
|
|
chr3_-_49823941
Show fit
|
0.08 |
ENST00000321599.4
ENST00000395238.1
ENST00000468463.1
ENST00000460540.1
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr19_+_10828724
Show fit
|
0.08 |
ENST00000585892.1
ENST00000314646.5
ENST00000359692.6
|
DNM2
|
dynamin 2
|
|
chr16_+_56782118
Show fit
|
0.08 |
ENST00000566678.1
|
NUP93
|
nucleoporin 93kDa
|
|
chr22_-_30234218
Show fit
|
0.08 |
ENST00000307790.3
ENST00000542393.1
ENST00000397771.2
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2
|
|
chr21_+_44394742
Show fit
|
0.08 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr19_-_51527467
Show fit
|
0.08 |
ENST00000593681.1
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr2_-_18770812
Show fit
|
0.08 |
ENST00000359846.2
ENST00000304081.4
ENST00000600945.1
ENST00000532967.1
ENST00000444297.2
|
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB
NT5C1B-RDH14 readthrough
|
|
chr1_+_228870824
Show fit
|
0.08 |
ENST00000366691.3
|
RHOU
|
ras homolog family member U
|
|
chr12_+_8850277
Show fit
|
0.08 |
ENST00000539923.1
ENST00000537189.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chrX_+_118425471
Show fit
|
0.08 |
ENST00000428222.1
|
RP5-1139I1.1
|
RP5-1139I1.1
|
|
chr13_-_34250861
Show fit
|
0.08 |
ENST00000445227.1
ENST00000454681.2
|
RP11-141M1.3
|
RP11-141M1.3
|
|
chr2_+_58134756
Show fit
|
0.08 |
ENST00000435505.2
ENST00000417641.2
|
VRK2
|
vaccinia related kinase 2
|
|
chr10_+_104005272
Show fit
|
0.08 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr2_+_62132800
Show fit
|
0.08 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1
|