Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
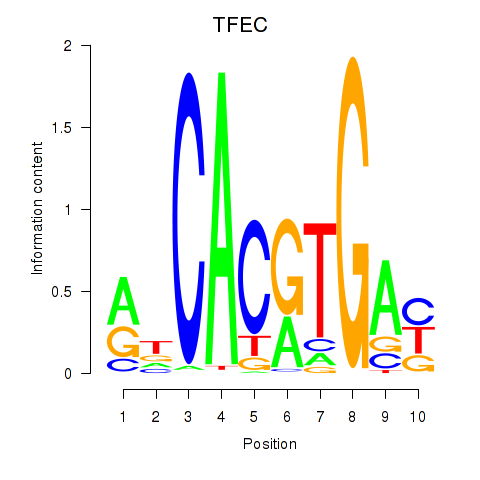
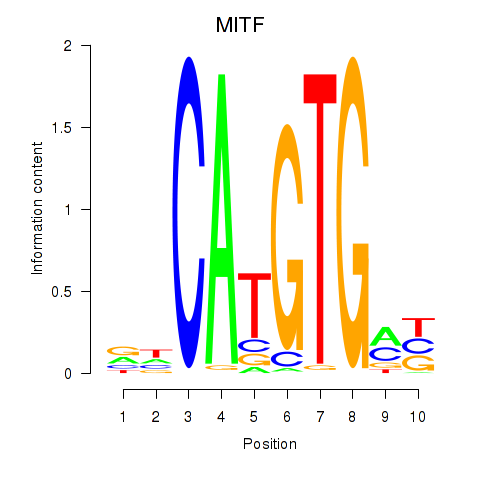
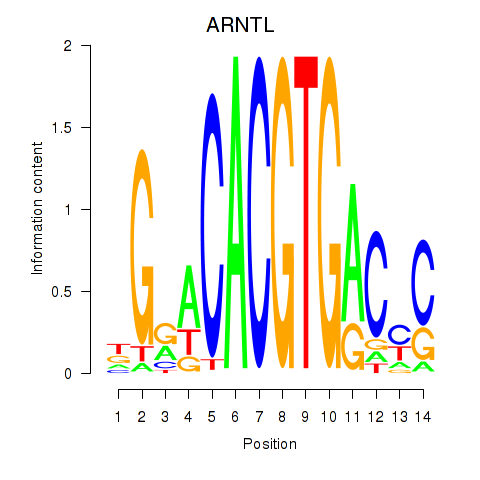
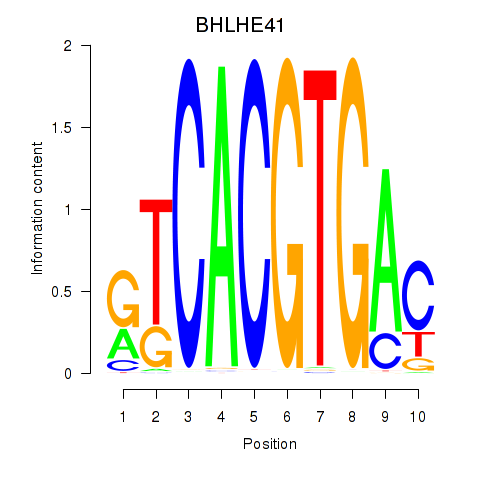
Results for TFEC_MITF_ARNTL_BHLHE41
Z-value: 1.20
Transcription factors associated with TFEC_MITF_ARNTL_BHLHE41
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFEC
|
ENSG00000105967.11 | transcription factor EC |
|
MITF
|
ENSG00000187098.10 | melanocyte inducing transcription factor |
|
ARNTL
|
ENSG00000133794.13 | aryl hydrocarbon receptor nuclear translocator like |
|
BHLHE41
|
ENSG00000123095.5 | basic helix-loop-helix family member e41 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE41 | hg19_v2_chr12_-_26278030_26278060 | 0.58 | 2.3e-01 | Click! |
| MITF | hg19_v2_chr3_+_69811858_69811881 | 0.57 | 2.4e-01 | Click! |
| TFEC | hg19_v2_chr7_-_115608304_115608352 | 0.53 | 2.7e-01 | Click! |
| ARNTL | hg19_v2_chr11_+_13299186_13299432 | 0.40 | 4.3e-01 | Click! |
Activity profile of TFEC_MITF_ARNTL_BHLHE41 motif
Sorted Z-values of TFEC_MITF_ARNTL_BHLHE41 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_109703663 | 1.56 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr10_-_46089939 | 1.52 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr6_-_33385854 | 1.39 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr19_+_5681011 | 1.37 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr13_+_113951607 | 1.37 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr11_+_67159416 | 1.15 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr1_-_183604794 | 1.13 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr2_-_176867534 | 0.93 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr6_-_84937314 | 0.93 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr8_+_103876528 | 0.93 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr19_-_5680891 | 0.89 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr10_-_46090334 | 0.88 |
ENST00000395771.3
ENST00000319836.3 |
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr19_+_5681153 | 0.87 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr8_-_54755459 | 0.86 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr16_+_28986134 | 0.86 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr5_+_133706865 | 0.85 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr19_+_1275917 | 0.85 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr1_+_42921761 | 0.83 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr8_-_71157595 | 0.81 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr17_-_7137582 | 0.79 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr5_+_41904431 | 0.77 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr17_+_42422629 | 0.76 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr18_+_9136758 | 0.75 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr4_-_47465666 | 0.74 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr15_-_72668805 | 0.72 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chrX_-_119693745 | 0.72 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr1_+_42922173 | 0.71 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr8_+_126103921 | 0.71 |
ENST00000523741.1
|
NSMCE2
|
non-SMC element 2, MMS21 homolog (S. cerevisiae) |
| chr14_-_54955721 | 0.70 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr17_-_7137857 | 0.70 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr16_+_28986085 | 0.68 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr5_+_110074685 | 0.68 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr19_+_10812108 | 0.68 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr6_+_87865262 | 0.66 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr3_-_49395892 | 0.66 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr12_+_56110247 | 0.65 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr13_+_113951532 | 0.65 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr3_-_49395705 | 0.65 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr12_+_56109810 | 0.60 |
ENST00000550412.1
ENST00000257899.2 ENST00000548925.1 ENST00000549147.1 |
RP11-644F5.10
BLOC1S1
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr7_+_99746514 | 0.60 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr16_-_1525016 | 0.59 |
ENST00000262318.8
ENST00000448525.1 |
CLCN7
|
chloride channel, voltage-sensitive 7 |
| chr1_-_154193091 | 0.58 |
ENST00000362076.4
ENST00000350592.3 ENST00000368516.1 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr7_+_150758642 | 0.58 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_-_18343725 | 0.58 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr16_-_5083917 | 0.58 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr3_-_113465065 | 0.58 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_+_56109926 | 0.58 |
ENST00000547076.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr8_-_54755789 | 0.57 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_-_68962744 | 0.56 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr10_+_70883908 | 0.56 |
ENST00000263559.6
ENST00000395098.1 ENST00000546041.1 ENST00000541711.1 |
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe) |
| chr3_-_122512619 | 0.53 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr11_-_71814422 | 0.53 |
ENST00000278671.5
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr22_-_36903101 | 0.52 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_-_71814276 | 0.51 |
ENST00000538404.1
ENST00000535107.1 ENST00000545249.1 |
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr19_-_5340730 | 0.51 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr6_-_109703634 | 0.50 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr14_-_81687197 | 0.50 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr17_-_76124812 | 0.49 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr17_+_42422662 | 0.49 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr16_+_2570340 | 0.48 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr4_+_76439095 | 0.48 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr1_-_68962782 | 0.47 |
ENST00000456315.2
|
DEPDC1
|
DEP domain containing 1 |
| chrX_+_150565038 | 0.47 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr6_-_33385902 | 0.47 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_1627834 | 0.46 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr12_-_76953513 | 0.46 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_-_54955376 | 0.46 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr17_+_78075361 | 0.46 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr1_-_33642151 | 0.45 |
ENST00000543586.1
|
TRIM62
|
tripartite motif containing 62 |
| chr22_-_42343117 | 0.45 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr10_+_99258625 | 0.45 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr22_-_36903069 | 0.45 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_+_42422637 | 0.44 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr4_+_76439665 | 0.43 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr8_-_145688231 | 0.43 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr17_-_79791118 | 0.43 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr2_-_40006289 | 0.42 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr5_-_176730733 | 0.42 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chrX_+_102585124 | 0.42 |
ENST00000332431.4
ENST00000372666.1 |
TCEAL7
|
transcription elongation factor A (SII)-like 7 |
| chrX_-_45629661 | 0.42 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr11_+_107879459 | 0.42 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr19_-_1237990 | 0.42 |
ENST00000382477.2
ENST00000215376.6 ENST00000590083.1 |
C19orf26
|
chromosome 19 open reading frame 26 |
| chr7_+_4815238 | 0.42 |
ENST00000348624.4
ENST00000401897.1 |
AP5Z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chrX_-_34675391 | 0.41 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr17_+_78075324 | 0.41 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chr2_-_40006357 | 0.40 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr19_-_49137762 | 0.40 |
ENST00000593500.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr16_-_4466565 | 0.40 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr19_+_41305330 | 0.40 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr11_-_6640585 | 0.39 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr7_+_100464760 | 0.39 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr13_+_42846272 | 0.38 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr2_+_240323439 | 0.38 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr17_+_62223320 | 0.37 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr6_-_109703600 | 0.37 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr12_-_76953284 | 0.37 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr11_-_1785139 | 0.36 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr1_-_222886526 | 0.36 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr17_+_78075498 | 0.36 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr15_-_76352069 | 0.36 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr11_+_65779283 | 0.36 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr8_-_82754427 | 0.35 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr22_-_42342692 | 0.35 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr19_-_36545128 | 0.35 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr13_+_49684445 | 0.35 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr19_-_49137790 | 0.35 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr3_-_196987309 | 0.35 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr17_+_6915730 | 0.34 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr5_+_43602750 | 0.34 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr12_+_28343353 | 0.34 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr17_+_6915798 | 0.34 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr9_-_90589586 | 0.34 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr19_+_8386371 | 0.34 |
ENST00000600659.2
|
RPS28
|
ribosomal protein S28 |
| chr22_-_50699972 | 0.33 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr18_-_29522989 | 0.33 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr11_-_18343669 | 0.32 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr4_+_56719782 | 0.32 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr4_-_23891693 | 0.31 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr9_-_90589402 | 0.31 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr6_-_33385870 | 0.31 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr7_+_150758304 | 0.31 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr2_+_109204909 | 0.31 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_-_103115135 | 0.31 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr7_+_99070527 | 0.31 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr2_+_46926326 | 0.30 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_-_47572207 | 0.30 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr19_-_5680499 | 0.30 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr22_-_39268192 | 0.30 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr19_-_5720123 | 0.30 |
ENST00000587365.1
ENST00000585374.1 ENST00000593119.1 |
LONP1
|
lon peptidase 1, mitochondrial |
| chr6_+_139349817 | 0.30 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr10_-_98031265 | 0.30 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr16_-_5083589 | 0.30 |
ENST00000563578.1
ENST00000562346.2 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr7_+_150759634 | 0.29 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr12_-_108154705 | 0.29 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr3_-_20227619 | 0.29 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr1_-_68962805 | 0.29 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr16_-_4466622 | 0.29 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr21_+_45287112 | 0.29 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr9_-_125667494 | 0.29 |
ENST00000335387.5
ENST00000357244.2 ENST00000373665.2 |
RC3H2
|
ring finger and CCCH-type domains 2 |
| chr14_-_54908043 | 0.28 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr15_-_50978965 | 0.27 |
ENST00000560955.1
ENST00000313478.7 |
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_+_74903580 | 0.27 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr7_+_116593536 | 0.27 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr3_-_134204815 | 0.27 |
ENST00000514612.1
ENST00000510994.1 ENST00000354910.5 |
ANAPC13
|
anaphase promoting complex subunit 13 |
| chr5_-_1799864 | 0.27 |
ENST00000510999.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr10_-_98031310 | 0.27 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr13_+_36920569 | 0.27 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr19_-_40854281 | 0.27 |
ENST00000392035.2
|
C19orf47
|
chromosome 19 open reading frame 47 |
| chr12_+_53645870 | 0.27 |
ENST00000329548.4
|
MFSD5
|
major facilitator superfamily domain containing 5 |
| chr8_+_109455830 | 0.27 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr19_-_5719860 | 0.27 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr7_+_23286182 | 0.27 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr2_+_46926048 | 0.27 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr19_-_8386238 | 0.27 |
ENST00000301457.2
|
NDUFA7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr16_+_2570431 | 0.27 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr1_+_61869748 | 0.27 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr16_-_70719925 | 0.26 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr2_-_148778323 | 0.26 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr19_-_5720248 | 0.26 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr4_-_54930790 | 0.26 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr6_-_80657292 | 0.26 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr12_+_56110315 | 0.26 |
ENST00000548556.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr17_-_18266818 | 0.26 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_-_148778258 | 0.25 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_+_100271446 | 0.25 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr3_-_120068143 | 0.25 |
ENST00000295628.3
|
LRRC58
|
leucine rich repeat containing 58 |
| chr2_+_172864490 | 0.25 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr12_+_28343365 | 0.25 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr19_+_797443 | 0.25 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr4_+_103790462 | 0.25 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chrX_+_102469997 | 0.25 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr2_+_228190066 | 0.25 |
ENST00000436237.1
ENST00000443428.2 ENST00000418961.1 |
MFF
|
mitochondrial fission factor |
| chr6_+_7590413 | 0.25 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr21_-_46237883 | 0.25 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr2_-_183903133 | 0.25 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr8_+_26240414 | 0.24 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr6_-_43197189 | 0.24 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr15_-_65282045 | 0.24 |
ENST00000558765.1
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr7_+_100271355 | 0.24 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr22_+_21996549 | 0.24 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr18_+_77867177 | 0.24 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr10_-_13342051 | 0.24 |
ENST00000479604.1
|
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr16_+_57139933 | 0.24 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr11_+_95523823 | 0.24 |
ENST00000538658.1
|
CEP57
|
centrosomal protein 57kDa |
| chr1_-_156265438 | 0.23 |
ENST00000362007.1
|
C1orf85
|
chromosome 1 open reading frame 85 |
| chr3_+_51428704 | 0.23 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr19_+_40697514 | 0.23 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr13_-_21635631 | 0.23 |
ENST00000382592.4
|
LATS2
|
large tumor suppressor kinase 2 |
| chr4_+_38665810 | 0.23 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr3_-_45883558 | 0.23 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_+_17248418 | 0.23 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr20_-_2821271 | 0.23 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr13_-_79233314 | 0.23 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr9_+_131084846 | 0.23 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr16_+_83986827 | 0.22 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr17_-_76124711 | 0.22 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr8_-_103876340 | 0.22 |
ENST00000518353.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr2_+_241526126 | 0.22 |
ENST00000391984.2
ENST00000391982.2 ENST00000404753.3 ENST00000270364.7 ENST00000352879.4 ENST00000354082.4 |
CAPN10
|
calpain 10 |
| chr19_-_11688260 | 0.22 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr10_+_69644404 | 0.22 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFEC_MITF_ARNTL_BHLHE41
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.4 | 2.0 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 1.5 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.3 | 0.8 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.3 | 0.8 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.3 | 1.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 0.8 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.2 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 1.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 0.7 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 2.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 1.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.3 | GO:1990828 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.1 | 0.6 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 1.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 1.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.3 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.2 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) regulation of protein localization to cilium(GO:1903564) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.5 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.2 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.2 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.5 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.4 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 2.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.6 | GO:0061008 | liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 1.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0006771 | riboflavin metabolic process(GO:0006771) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.2 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0060592 | mammary gland formation(GO:0060592) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 1.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 0.9 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.3 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.2 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 2.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0072589 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 1.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.3 | 0.8 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.2 | 0.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 1.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.5 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 0.8 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 1.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.1 | 0.2 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.2 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.2 | GO:0031177 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.2 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.4 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |