Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
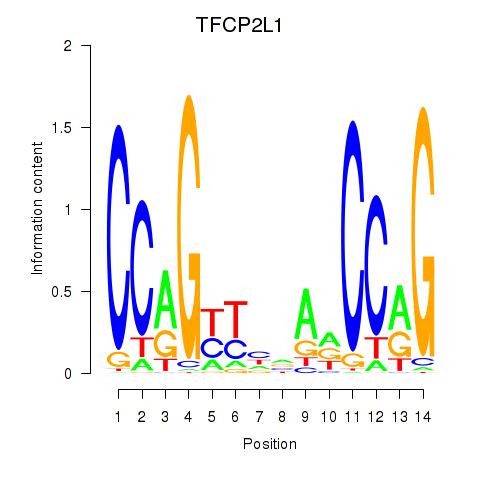
Results for TFCP2L1
Z-value: 0.41
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.14 | 7.9e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106069247 | 0.27 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chr15_-_90233907 | 0.20 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr14_+_64971438 | 0.19 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr18_-_59415987 | 0.15 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr19_+_41860047 | 0.15 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr19_-_12405689 | 0.15 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr9_-_117160738 | 0.15 |
ENST00000448674.1
|
RP11-9M16.2
|
RP11-9M16.2 |
| chr11_+_65265141 | 0.13 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr2_+_95873257 | 0.12 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr11_+_1856034 | 0.10 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr8_-_54755459 | 0.10 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr19_-_42916499 | 0.09 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr2_-_61389240 | 0.09 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr17_+_38337491 | 0.09 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_-_89458287 | 0.09 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr7_+_64363625 | 0.09 |
ENST00000476120.1
ENST00000319636.5 ENST00000545510.1 |
ZNF273
|
zinc finger protein 273 |
| chr19_+_11877838 | 0.09 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr6_-_30710265 | 0.09 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr2_+_17997763 | 0.09 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr19_-_12476443 | 0.08 |
ENST00000242804.4
ENST00000438182.1 |
ZNF442
|
zinc finger protein 442 |
| chr11_-_119293872 | 0.08 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr11_-_76155700 | 0.08 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_27668505 | 0.08 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr7_-_99756293 | 0.08 |
ENST00000316937.3
ENST00000456769.1 |
C7orf43
|
chromosome 7 open reading frame 43 |
| chr20_+_44098346 | 0.08 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr6_-_52859968 | 0.07 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr11_+_17298297 | 0.07 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr12_+_82752647 | 0.07 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chrX_+_150565653 | 0.07 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr2_+_219125714 | 0.07 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr6_-_52860171 | 0.07 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr11_+_17298255 | 0.07 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr19_+_12721760 | 0.07 |
ENST00000600752.1
ENST00000540038.1 |
ZNF791
|
zinc finger protein 791 |
| chr11_+_392587 | 0.07 |
ENST00000534401.1
|
PKP3
|
plakophilin 3 |
| chr5_+_162864575 | 0.06 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr17_+_62503073 | 0.06 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr1_-_89458604 | 0.06 |
ENST00000260508.4
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr1_-_139379 | 0.06 |
ENST00000423372.3
|
AL627309.1
|
Uncharacterized protein |
| chr9_+_88556036 | 0.06 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chrX_+_49687267 | 0.06 |
ENST00000376091.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chrX_+_24167828 | 0.06 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr5_-_148929848 | 0.06 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr22_+_25595817 | 0.06 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr3_-_122134882 | 0.06 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr5_+_79703823 | 0.06 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr7_+_63774321 | 0.06 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr19_-_20150252 | 0.06 |
ENST00000595736.1
ENST00000593468.1 ENST00000596019.1 |
ZNF682
|
zinc finger protein 682 |
| chrX_+_49687216 | 0.06 |
ENST00000376088.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr5_+_150051149 | 0.06 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr2_-_20212422 | 0.06 |
ENST00000421259.2
ENST00000407540.3 |
MATN3
|
matrilin 3 |
| chr12_-_110883346 | 0.06 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr12_-_74686314 | 0.06 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr3_-_123411191 | 0.05 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr12_+_82752283 | 0.05 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr8_+_117778736 | 0.05 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr17_-_66287257 | 0.05 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr9_-_130829588 | 0.05 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr4_-_122744998 | 0.05 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr1_+_174843548 | 0.05 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr10_-_61469837 | 0.05 |
ENST00000395348.3
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr18_-_2982869 | 0.05 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr1_+_110198689 | 0.05 |
ENST00000369836.4
|
GSTM4
|
glutathione S-transferase mu 4 |
| chr6_-_30709980 | 0.05 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr22_+_50986462 | 0.05 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr20_+_30458431 | 0.05 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr4_-_57301748 | 0.05 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr12_-_66563786 | 0.05 |
ENST00000542724.1
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr5_+_34685805 | 0.05 |
ENST00000508315.1
|
RAI14
|
retinoic acid induced 14 |
| chr3_-_50605077 | 0.05 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr17_+_66287628 | 0.05 |
ENST00000581639.1
ENST00000452479.2 |
ARSG
|
arylsulfatase G |
| chr17_+_62503147 | 0.05 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr6_+_149068464 | 0.05 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr19_-_20844343 | 0.05 |
ENST00000595405.1
|
ZNF626
|
zinc finger protein 626 |
| chr19_-_20150014 | 0.05 |
ENST00000358523.5
ENST00000397162.1 ENST00000601100.1 |
ZNF682
|
zinc finger protein 682 |
| chr19_-_12662314 | 0.05 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr11_+_62649158 | 0.05 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr12_-_124456598 | 0.05 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chrX_-_106960285 | 0.05 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr11_-_46848393 | 0.04 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr1_-_155959280 | 0.04 |
ENST00000495070.2
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_49170405 | 0.04 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr13_-_36705425 | 0.04 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr8_-_103876965 | 0.04 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr1_-_98386543 | 0.04 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr6_-_31080336 | 0.04 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr22_-_39151434 | 0.04 |
ENST00000439339.1
|
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr7_-_64467031 | 0.04 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr1_+_110198714 | 0.04 |
ENST00000336075.5
ENST00000326729.5 |
GSTM4
|
glutathione S-transferase mu 4 |
| chr1_-_113161730 | 0.04 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr16_+_22019404 | 0.04 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr10_+_35416223 | 0.04 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr4_-_25865159 | 0.04 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_+_110693103 | 0.04 |
ENST00000331565.4
|
SLC6A17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr8_+_103876528 | 0.04 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr19_-_12251130 | 0.04 |
ENST00000418866.1
ENST00000600335.1 |
ZNF20
|
zinc finger protein 20 |
| chr9_-_129885010 | 0.04 |
ENST00000373425.3
|
ANGPTL2
|
angiopoietin-like 2 |
| chr11_+_17298522 | 0.04 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr19_-_12405606 | 0.04 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chrX_-_63425561 | 0.04 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr1_+_55107449 | 0.04 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr17_-_27278445 | 0.04 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr1_+_205225319 | 0.04 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr20_+_34680595 | 0.04 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr13_-_20806440 | 0.04 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr1_-_113162040 | 0.04 |
ENST00000358039.4
ENST00000369668.2 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr8_+_73921085 | 0.04 |
ENST00000276603.5
ENST00000276602.6 ENST00000518874.1 |
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr10_+_99344071 | 0.04 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr10_+_72972281 | 0.04 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr22_-_39150947 | 0.04 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr12_-_48500085 | 0.04 |
ENST00000549518.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr5_-_131826457 | 0.04 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr6_+_84569359 | 0.04 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr8_-_124553437 | 0.04 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr19_+_38880695 | 0.04 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_+_17298543 | 0.04 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chrX_-_138724677 | 0.03 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_+_65190245 | 0.03 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr19_+_41305612 | 0.03 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_-_48499826 | 0.03 |
ENST00000551798.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr17_-_73266616 | 0.03 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr3_-_52322019 | 0.03 |
ENST00000463624.1
|
WDR82
|
WD repeat domain 82 |
| chr21_-_16437126 | 0.03 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr1_-_156217829 | 0.03 |
ENST00000356983.2
ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr7_-_64023441 | 0.03 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr12_+_14572070 | 0.03 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr3_-_148804275 | 0.03 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr19_+_41305330 | 0.03 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_23299777 | 0.03 |
ENST00000597761.2
|
ZNF730
|
zinc finger protein 730 |
| chr17_-_66287350 | 0.03 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr11_+_77899842 | 0.03 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr12_+_7941989 | 0.03 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr12_-_48499591 | 0.03 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr14_-_61190754 | 0.03 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr17_-_27467418 | 0.03 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr17_+_40996590 | 0.03 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr1_-_156217822 | 0.03 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr3_+_38323785 | 0.03 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr7_+_77167376 | 0.03 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr9_-_117150303 | 0.03 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chrX_-_51239425 | 0.03 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr2_-_233415220 | 0.03 |
ENST00000408957.3
|
TIGD1
|
tigger transposable element derived 1 |
| chr7_+_6655225 | 0.03 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr11_+_67776012 | 0.03 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr7_+_43966018 | 0.03 |
ENST00000222402.3
ENST00000446008.1 ENST00000394798.4 |
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr3_+_184081137 | 0.03 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_153538292 | 0.03 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr3_-_52488048 | 0.03 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr19_+_44100544 | 0.03 |
ENST00000391965.2
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr17_+_26833250 | 0.03 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr4_-_987164 | 0.03 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr19_+_41305627 | 0.03 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_11750566 | 0.03 |
ENST00000344893.3
|
ZNF833P
|
zinc finger protein 833, pseudogene |
| chr6_+_31554962 | 0.03 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr12_-_120241187 | 0.03 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr19_+_55795493 | 0.03 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr1_-_43751230 | 0.03 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr5_-_180018540 | 0.03 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr4_+_57302297 | 0.03 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_-_55686399 | 0.02 |
ENST00000587067.1
|
SYT5
|
synaptotagmin V |
| chr19_+_12035878 | 0.02 |
ENST00000254321.5
ENST00000538752.1 ENST00000590798.1 |
ZNF700
ZNF763
ZNF763
|
zinc finger protein 700 zinc finger protein 763 Uncharacterized protein; Zinc finger protein 763 |
| chr2_+_241544834 | 0.02 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr9_+_101867387 | 0.02 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr17_+_55183261 | 0.02 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr12_-_121712313 | 0.02 |
ENST00000392474.2
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr11_+_96123158 | 0.02 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr9_-_129884902 | 0.02 |
ENST00000373417.1
|
ANGPTL2
|
angiopoietin-like 2 |
| chr16_+_57023406 | 0.02 |
ENST00000262510.6
ENST00000308149.7 ENST00000436936.1 |
NLRC5
|
NLR family, CARD domain containing 5 |
| chr15_-_64665911 | 0.02 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr22_-_39151463 | 0.02 |
ENST00000405510.1
ENST00000433561.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr5_-_58652788 | 0.02 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr14_+_32546274 | 0.02 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr14_-_64971288 | 0.02 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr20_+_44098385 | 0.02 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr11_-_123612319 | 0.02 |
ENST00000526252.1
ENST00000530393.1 ENST00000533463.1 ENST00000336139.4 ENST00000529691.1 ENST00000528306.1 |
ZNF202
|
zinc finger protein 202 |
| chr21_-_16437255 | 0.02 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr11_+_77899920 | 0.02 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr1_+_116654376 | 0.02 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr22_+_21369316 | 0.02 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr2_+_130737223 | 0.02 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr12_+_52695617 | 0.02 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr3_-_50605150 | 0.02 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr11_+_133902547 | 0.02 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr1_-_89458415 | 0.02 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr15_-_62352570 | 0.02 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr11_-_62380199 | 0.02 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr12_+_124118366 | 0.02 |
ENST00000539994.1
ENST00000538845.1 ENST00000228955.7 ENST00000543341.2 ENST00000536375.1 |
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa |
| chr4_+_57301896 | 0.02 |
ENST00000514888.1
ENST00000264221.2 ENST00000505164.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr9_+_71394945 | 0.02 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr14_-_61124977 | 0.02 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr3_-_176915215 | 0.02 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr6_+_30978251 | 0.02 |
ENST00000561890.1
|
MUC22
|
mucin 22 |
| chr11_-_18343725 | 0.02 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr1_+_113161778 | 0.02 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr3_+_69915363 | 0.02 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_85780086 | 0.02 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_226129189 | 0.02 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr19_-_12721616 | 0.02 |
ENST00000311437.6
|
ZNF490
|
zinc finger protein 490 |
| chr11_-_96123022 | 0.02 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr18_-_24765248 | 0.02 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr11_-_65374430 | 0.02 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.0 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |