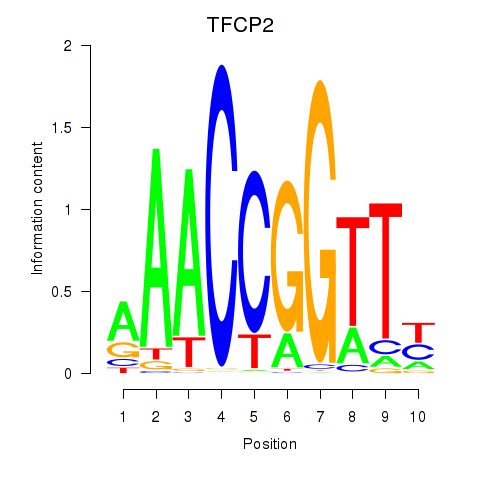
|
chr2_-_61765315
Show fit
|
1.14 |
ENST00000406957.1
ENST00000401558.2
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr4_+_26859300
Show fit
|
0.93 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2
|
|
chr2_-_61765732
Show fit
|
0.87 |
ENST00000443240.1
ENST00000436018.1
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr4_-_129207942
Show fit
|
0.82 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr14_-_71107921
Show fit
|
0.80 |
ENST00000553982.1
ENST00000500016.1
|
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5
CTD-2540L5.6
|
|
chrX_-_119693745
Show fit
|
0.79 |
ENST00000371323.2
|
CUL4B
|
cullin 4B
|
|
chr10_+_6625605
Show fit
|
0.77 |
ENST00000414894.1
ENST00000449648.1
|
PRKCQ-AS1
|
PRKCQ antisense RNA 1
|
|
chr1_+_206623784
Show fit
|
0.73 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr8_+_95731904
Show fit
|
0.63 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr1_-_145382434
Show fit
|
0.62 |
ENST00000610154.1
|
RP11-458D21.1
|
RP11-458D21.1
|
|
chr4_-_129208030
Show fit
|
0.57 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr11_-_85430088
Show fit
|
0.56 |
ENST00000533057.1
ENST00000533892.1
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_132362226
Show fit
|
0.55 |
ENST00000509437.1
ENST00000355372.2
ENST00000513541.1
ENST00000509008.1
ENST00000513848.1
ENST00000504170.1
ENST00000324170.3
|
ZCCHC10
|
zinc finger, CCHC domain containing 10
|
|
chr11_-_96123022
Show fit
|
0.55 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr8_-_95487272
Show fit
|
0.55 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr2_+_189156389
Show fit
|
0.55 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr14_-_68157084
Show fit
|
0.54 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4
|
|
chr14_+_32547434
Show fit
|
0.54 |
ENST00000556191.1
ENST00000554090.1
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr8_-_17533838
Show fit
|
0.53 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr11_+_65265141
Show fit
|
0.52 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr14_-_106069247
Show fit
|
0.52 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1
|
|
chr1_+_86889769
Show fit
|
0.52 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2
|
|
chr1_-_145382362
Show fit
|
0.51 |
ENST00000419817.1
ENST00000421937.3
ENST00000433081.2
|
RP11-458D21.1
|
RP11-458D21.1
|
|
chr1_-_221915418
Show fit
|
0.51 |
ENST00000323825.3
ENST00000366899.3
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr3_-_113464906
Show fit
|
0.50 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr2_-_55277692
Show fit
|
0.50 |
ENST00000394611.2
|
RTN4
|
reticulon 4
|
|
chr4_-_153274078
Show fit
|
0.49 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase
|
|
chr5_+_67584523
Show fit
|
0.49 |
ENST00000521409.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chrX_+_105937068
Show fit
|
0.49 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase
|
|
chr2_-_55277654
Show fit
|
0.47 |
ENST00000337526.6
ENST00000317610.7
ENST00000357732.4
|
RTN4
|
reticulon 4
|
|
chr12_+_72061563
Show fit
|
0.47 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr11_+_47270475
Show fit
|
0.47 |
ENST00000481889.2
ENST00000436778.1
ENST00000531660.1
ENST00000407404.1
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr7_-_120497178
Show fit
|
0.46 |
ENST00000441017.1
ENST00000424710.1
ENST00000433758.1
|
TSPAN12
|
tetraspanin 12
|
|
chr15_+_63682335
Show fit
|
0.46 |
ENST00000559379.1
ENST00000559821.1
|
RP11-321G12.1
|
RP11-321G12.1
|
|
chr6_-_84937314
Show fit
|
0.45 |
ENST00000257766.4
ENST00000403245.3
|
KIAA1009
|
KIAA1009
|
|
chr19_+_507299
Show fit
|
0.45 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr6_+_63921351
Show fit
|
0.44 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C
|
|
chr16_+_67193834
Show fit
|
0.43 |
ENST00000258200.3
ENST00000518148.1
ENST00000519917.1
ENST00000517382.1
ENST00000521920.1
|
FBXL8
|
F-box and leucine-rich repeat protein 8
|
|
chr2_+_172290707
Show fit
|
0.43 |
ENST00000375255.3
ENST00000539783.1
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr12_+_42725554
Show fit
|
0.42 |
ENST00000546750.1
ENST00000547847.1
|
PPHLN1
|
periphilin 1
|
|
chr22_-_31742218
Show fit
|
0.42 |
ENST00000266269.5
ENST00000405309.3
ENST00000351933.4
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr1_+_164528866
Show fit
|
0.42 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr18_+_61575200
Show fit
|
0.41 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr8_-_80680078
Show fit
|
0.41 |
ENST00000337919.5
ENST00000354724.3
|
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1
|
|
chr5_+_65222384
Show fit
|
0.41 |
ENST00000380943.2
ENST00000416865.2
ENST00000380939.2
ENST00000380936.1
ENST00000380935.1
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr11_-_85430163
Show fit
|
0.40 |
ENST00000529581.1
ENST00000533577.1
|
SYTL2
|
synaptotagmin-like 2
|
|
chr1_+_117963209
Show fit
|
0.40 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chr15_-_42565221
Show fit
|
0.39 |
ENST00000563371.1
ENST00000568400.1
ENST00000568432.1
|
TMEM87A
|
transmembrane protein 87A
|
|
chrX_+_105936982
Show fit
|
0.38 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase
|
|
chr15_-_42565023
Show fit
|
0.38 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A
|
|
chr17_+_7608511
Show fit
|
0.38 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3
|
|
chr10_+_64133934
Show fit
|
0.38 |
ENST00000395254.3
ENST00000395255.3
ENST00000410046.3
|
ZNF365
|
zinc finger protein 365
|
|
chr3_-_42845922
Show fit
|
0.38 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr19_-_11849697
Show fit
|
0.37 |
ENST00000586121.1
ENST00000431998.1
ENST00000341191.6
ENST00000545749.1
ENST00000440527.1
|
ZNF823
|
zinc finger protein 823
|
|
chr9_-_21995249
Show fit
|
0.37 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A
|
|
chr10_-_33625154
Show fit
|
0.37 |
ENST00000265371.4
|
NRP1
|
neuropilin 1
|
|
chr8_-_95487331
Show fit
|
0.37 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr1_+_87797351
Show fit
|
0.37 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4
|
|
chr5_+_65222500
Show fit
|
0.37 |
ENST00000511297.1
ENST00000506030.1
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr9_+_125703282
Show fit
|
0.36 |
ENST00000373647.4
ENST00000402311.1
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr5_+_136070614
Show fit
|
0.36 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1
|
|
chr12_-_118796910
Show fit
|
0.36 |
ENST00000541186.1
ENST00000539872.1
|
TAOK3
|
TAO kinase 3
|
|
chr2_+_238395042
Show fit
|
0.36 |
ENST00000429898.1
ENST00000410032.1
|
MLPH
|
melanophilin
|
|
chr2_+_238395879
Show fit
|
0.36 |
ENST00000445024.2
ENST00000338530.4
ENST00000409373.1
|
MLPH
|
melanophilin
|
|
chr3_+_108321623
Show fit
|
0.35 |
ENST00000497905.1
ENST00000463306.1
|
DZIP3
|
DAZ interacting zinc finger protein 3
|
|
chrX_+_119737806
Show fit
|
0.35 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr5_+_65222438
Show fit
|
0.34 |
ENST00000380938.2
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr2_-_55277512
Show fit
|
0.34 |
ENST00000402434.2
|
RTN4
|
reticulon 4
|
|
chr8_-_103250997
Show fit
|
0.34 |
ENST00000522368.1
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible)
|
|
chr16_-_20817753
Show fit
|
0.33 |
ENST00000389345.5
ENST00000300005.3
ENST00000357967.4
ENST00000569729.1
|
ERI2
|
ERI1 exoribonuclease family member 2
|
|
chr5_-_39219705
Show fit
|
0.33 |
ENST00000351578.6
|
FYB
|
FYN binding protein
|
|
chr3_-_113465065
Show fit
|
0.33 |
ENST00000497255.1
ENST00000478020.1
ENST00000240922.3
ENST00000493900.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr4_+_128702969
Show fit
|
0.33 |
ENST00000508776.1
ENST00000439123.2
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chr12_+_41086215
Show fit
|
0.33 |
ENST00000547702.1
ENST00000551424.1
|
CNTN1
|
contactin 1
|
|
chr1_-_236046872
Show fit
|
0.33 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator
|
|
chr6_+_142623758
Show fit
|
0.33 |
ENST00000541199.1
ENST00000435011.2
|
GPR126
|
G protein-coupled receptor 126
|
|
chr10_-_76995675
Show fit
|
0.32 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr6_+_142623063
Show fit
|
0.32 |
ENST00000296932.8
ENST00000367609.3
|
GPR126
|
G protein-coupled receptor 126
|
|
chr2_-_55277436
Show fit
|
0.32 |
ENST00000354474.6
|
RTN4
|
reticulon 4
|
|
chr6_-_73935163
Show fit
|
0.32 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like
|
|
chr1_-_229644034
Show fit
|
0.31 |
ENST00000366678.3
ENST00000261396.3
ENST00000537506.1
|
NUP133
|
nucleoporin 133kDa
|
|
chr17_-_42991062
Show fit
|
0.31 |
ENST00000587997.1
|
GFAP
|
glial fibrillary acidic protein
|
|
chr6_+_155470243
Show fit
|
0.31 |
ENST00000456877.2
ENST00000528391.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr19_-_57678811
Show fit
|
0.30 |
ENST00000554048.2
|
DUXA
|
double homeobox A
|
|
chr12_-_118796971
Show fit
|
0.30 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3
|
|
chr19_-_12146483
Show fit
|
0.30 |
ENST00000455504.2
ENST00000547560.1
ENST00000552904.1
ENST00000419886.2
ENST00000550507.1
ENST00000344980.6
ENST00000550745.1
ENST00000411841.1
|
ZNF433
|
zinc finger protein 433
|
|
chr2_-_152146385
Show fit
|
0.30 |
ENST00000414946.1
ENST00000243346.5
|
NMI
|
N-myc (and STAT) interactor
|
|
chr6_+_3000195
Show fit
|
0.29 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr22_+_24199044
Show fit
|
0.29 |
ENST00000403208.3
ENST00000398356.2
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11
|
|
chrX_+_123095890
Show fit
|
0.29 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2
|
|
chr17_-_5138099
Show fit
|
0.28 |
ENST00000571800.1
ENST00000574081.1
ENST00000399600.4
ENST00000574297.1
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chrX_-_72434628
Show fit
|
0.28 |
ENST00000536638.1
ENST00000373517.3
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr14_-_24685246
Show fit
|
0.28 |
ENST00000396833.2
ENST00000288087.7
|
MDP1
|
magnesium-dependent phosphatase 1
|
|
chr8_-_54755459
Show fit
|
0.28 |
ENST00000524234.1
ENST00000521275.1
ENST00000396774.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr20_-_5591626
Show fit
|
0.28 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr5_-_148930960
Show fit
|
0.28 |
ENST00000261798.5
ENST00000377843.2
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr1_-_27709816
Show fit
|
0.28 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2
|
|
chr12_+_57522692
Show fit
|
0.28 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr16_+_57653989
Show fit
|
0.28 |
ENST00000567835.1
ENST00000569372.1
ENST00000563548.1
ENST00000562003.1
|
GPR56
|
G protein-coupled receptor 56
|
|
chr2_-_71222466
Show fit
|
0.27 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein
|
|
chr1_-_85040090
Show fit
|
0.27 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl-
|
|
chrX_+_133507327
Show fit
|
0.27 |
ENST00000332070.3
ENST00000394292.1
ENST00000370799.1
ENST00000416404.2
|
PHF6
|
PHD finger protein 6
|
|
chr6_+_3000218
Show fit
|
0.27 |
ENST00000380441.1
ENST00000380455.4
ENST00000380454.4
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr1_-_54304212
Show fit
|
0.27 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin
|
|
chr5_-_39219641
Show fit
|
0.26 |
ENST00000509072.1
ENST00000504542.1
ENST00000505428.1
ENST00000506557.1
|
FYB
|
FYN binding protein
|
|
chr11_-_28129656
Show fit
|
0.26 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A
|
|
chr3_+_152552685
Show fit
|
0.26 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1
|
|
chr2_+_63277927
Show fit
|
0.26 |
ENST00000282549.2
|
OTX1
|
orthodenticle homeobox 1
|
|
chr1_-_39325431
Show fit
|
0.26 |
ENST00000373001.3
|
RRAGC
|
Ras-related GTP binding C
|
|
chr4_+_170581213
Show fit
|
0.26 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3
|
|
chr19_+_38826415
Show fit
|
0.25 |
ENST00000410018.1
ENST00000409235.3
|
CATSPERG
|
catsper channel auxiliary subunit gamma
|
|
chr20_-_46415297
Show fit
|
0.25 |
ENST00000467815.1
ENST00000359930.4
|
SULF2
|
sulfatase 2
|
|
chr5_-_111093406
Show fit
|
0.25 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein
|
|
chr2_+_103089756
Show fit
|
0.25 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4
|
|
chr5_-_111093167
Show fit
|
0.25 |
ENST00000446294.2
ENST00000419114.2
|
NREP
|
neuronal regeneration related protein
|
|
chr20_-_46415341
Show fit
|
0.24 |
ENST00000484875.1
ENST00000361612.4
|
SULF2
|
sulfatase 2
|
|
chr17_-_7082861
Show fit
|
0.24 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr2_-_188378368
Show fit
|
0.24 |
ENST00000392365.1
ENST00000435414.1
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_56150184
Show fit
|
0.24 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr18_-_54305658
Show fit
|
0.24 |
ENST00000586262.1
ENST00000217515.6
|
TXNL1
|
thioredoxin-like 1
|
|
chr16_+_78133293
Show fit
|
0.24 |
ENST00000566780.1
|
WWOX
|
WW domain containing oxidoreductase
|
|
chr8_-_28243934
Show fit
|
0.24 |
ENST00000521185.1
ENST00000520290.1
ENST00000344423.5
|
ZNF395
|
zinc finger protein 395
|
|
chr11_+_47270436
Show fit
|
0.24 |
ENST00000395397.3
ENST00000405576.1
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr12_-_76478446
Show fit
|
0.23 |
ENST00000393263.3
ENST00000548044.1
ENST00000547704.1
ENST00000431879.3
ENST00000549596.1
ENST00000550934.1
ENST00000551600.1
ENST00000547479.1
ENST00000547773.1
ENST00000544816.1
ENST00000542344.1
ENST00000548273.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr20_+_11873141
Show fit
|
0.23 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr19_-_37019136
Show fit
|
0.23 |
ENST00000592282.1
|
ZNF260
|
zinc finger protein 260
|
|
chr11_-_18956556
Show fit
|
0.23 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1
|
|
chr14_+_75746781
Show fit
|
0.23 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr20_+_34042962
Show fit
|
0.23 |
ENST00000446710.1
ENST00000420564.1
|
CEP250
|
centrosomal protein 250kDa
|
|
chr6_+_64282447
Show fit
|
0.22 |
ENST00000370650.2
ENST00000578299.1
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr5_-_148930731
Show fit
|
0.22 |
ENST00000515748.2
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr5_+_65222299
Show fit
|
0.22 |
ENST00000284037.5
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr22_-_37571089
Show fit
|
0.22 |
ENST00000453962.1
ENST00000429622.1
ENST00000445595.1
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr1_-_67600639
Show fit
|
0.22 |
ENST00000544837.1
ENST00000603691.1
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr6_-_8102714
Show fit
|
0.21 |
ENST00000502429.1
ENST00000429723.2
ENST00000507463.1
ENST00000379715.5
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1
|
|
chr12_+_41086297
Show fit
|
0.21 |
ENST00000551295.2
|
CNTN1
|
contactin 1
|
|
chr19_-_3600549
Show fit
|
0.21 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor
|
|
chr4_-_99850243
Show fit
|
0.20 |
ENST00000280892.6
ENST00000511644.1
ENST00000504432.1
ENST00000505992.1
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr5_+_32711829
Show fit
|
0.20 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr11_+_114271251
Show fit
|
0.20 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7
|
|
chr8_-_36636676
Show fit
|
0.20 |
ENST00000524132.1
ENST00000519451.1
|
RP11-962G15.1
|
RP11-962G15.1
|
|
chrX_+_133507283
Show fit
|
0.20 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6
|
|
chr8_+_120428546
Show fit
|
0.20 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed
|
|
chrX_+_133941250
Show fit
|
0.20 |
ENST00000445123.1
|
FAM122C
|
family with sequence similarity 122C
|
|
chr11_+_62649158
Show fit
|
0.20 |
ENST00000539891.1
ENST00000536981.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2
|
|
chr7_+_127233689
Show fit
|
0.20 |
ENST00000265825.5
ENST00000420086.2
|
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus)
|
|
chr11_+_134225909
Show fit
|
0.19 |
ENST00000533324.1
|
GLB1L2
|
galactosidase, beta 1-like 2
|
|
chr11_+_28129795
Show fit
|
0.19 |
ENST00000406787.3
ENST00000342303.5
ENST00000403099.1
ENST00000407364.3
|
METTL15
|
methyltransferase like 15
|
|
chr5_-_111092873
Show fit
|
0.19 |
ENST00000509025.1
ENST00000515855.1
|
NREP
|
neuronal regeneration related protein
|
|
chr16_-_82045049
Show fit
|
0.19 |
ENST00000532128.1
ENST00000328945.5
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr2_+_238395803
Show fit
|
0.19 |
ENST00000264605.3
|
MLPH
|
melanophilin
|
|
chr6_-_30181133
Show fit
|
0.19 |
ENST00000454678.2
ENST00000434785.1
|
TRIM26
|
tripartite motif containing 26
|
|
chr3_+_107241783
Show fit
|
0.19 |
ENST00000415149.2
ENST00000402543.1
ENST00000325805.8
ENST00000427402.1
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr1_+_160321120
Show fit
|
0.19 |
ENST00000424754.1
|
NCSTN
|
nicastrin
|
|
chr4_-_157892055
Show fit
|
0.19 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C
|
|
chr11_+_114271314
Show fit
|
0.19 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7
|
|
chr5_-_37371163
Show fit
|
0.19 |
ENST00000513532.1
|
NUP155
|
nucleoporin 155kDa
|
|
chr12_+_6561190
Show fit
|
0.18 |
ENST00000544021.1
ENST00000266556.7
|
TAPBPL
|
TAP binding protein-like
|
|
chr2_+_240684554
Show fit
|
0.18 |
ENST00000407524.1
|
AC093802.1
|
Uncharacterized protein
|
|
chr2_-_85555355
Show fit
|
0.18 |
ENST00000282120.2
ENST00000398263.2
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr14_+_74815116
Show fit
|
0.18 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated
|
|
chr14_-_82089405
Show fit
|
0.18 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1
|
|
chr5_+_110427983
Show fit
|
0.18 |
ENST00000513710.2
ENST00000505303.1
|
WDR36
|
WD repeat domain 36
|
|
chr14_-_23451467
Show fit
|
0.17 |
ENST00000555074.1
ENST00000361265.4
|
RP11-298I3.5
AJUBA
|
RP11-298I3.5
ajuba LIM protein
|
|
chrX_-_46759138
Show fit
|
0.17 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31
|
|
chr4_-_107237340
Show fit
|
0.17 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase
|
|
chrX_-_118925600
Show fit
|
0.17 |
ENST00000361575.3
|
RPL39
|
ribosomal protein L39
|
|
chr17_-_7137582
Show fit
|
0.17 |
ENST00000575756.1
ENST00000575458.1
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr4_+_166794383
Show fit
|
0.17 |
ENST00000061240.2
ENST00000507499.1
|
TLL1
|
tolloid-like 1
|
|
chr5_+_54455946
Show fit
|
0.17 |
ENST00000503787.1
ENST00000296734.6
ENST00000515370.1
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr2_+_200820269
Show fit
|
0.17 |
ENST00000392290.1
|
C2orf47
|
chromosome 2 open reading frame 47
|
|
chr5_-_36242119
Show fit
|
0.17 |
ENST00000511088.1
ENST00000282512.3
ENST00000506945.1
|
NADK2
|
NAD kinase 2, mitochondrial
|
|
chr5_+_140782351
Show fit
|
0.17 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr13_+_52586517
Show fit
|
0.16 |
ENST00000523764.1
ENST00000521508.1
|
ALG11
|
ALG11, alpha-1,2-mannosyltransferase
|
|
chr17_+_38474489
Show fit
|
0.16 |
ENST00000394089.2
ENST00000425707.3
|
RARA
|
retinoic acid receptor, alpha
|
|
chr4_-_107237374
Show fit
|
0.16 |
ENST00000361687.4
ENST00000507696.1
ENST00000394708.2
ENST00000509532.1
|
TBCK
|
TBC1 domain containing kinase
|
|
chr22_+_45898712
Show fit
|
0.16 |
ENST00000455233.1
ENST00000348697.2
ENST00000402984.3
ENST00000262722.7
ENST00000327858.6
ENST00000442170.2
ENST00000340923.5
ENST00000439835.1
|
FBLN1
|
fibulin 1
|
|
chr16_-_3545424
Show fit
|
0.16 |
ENST00000437192.3
ENST00000399645.3
|
C16orf90
|
chromosome 16 open reading frame 90
|
|
chr2_+_191792376
Show fit
|
0.16 |
ENST00000409428.1
ENST00000409215.1
|
GLS
|
glutaminase
|
|
chr11_+_31391381
Show fit
|
0.16 |
ENST00000465995.1
ENST00000536040.1
|
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24
|
|
chr4_-_174255536
Show fit
|
0.16 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2
|
|
chr22_+_24199113
Show fit
|
0.15 |
ENST00000405847.1
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11
|
|
chr5_-_37371278
Show fit
|
0.15 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa
|
|
chr3_-_142297668
Show fit
|
0.15 |
ENST00000350721.4
ENST00000383101.3
|
ATR
|
ataxia telangiectasia and Rad3 related
|
|
chr17_-_33905521
Show fit
|
0.15 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12
|
|
chr1_+_45212051
Show fit
|
0.15 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C
|
|
chr19_+_38826477
Show fit
|
0.15 |
ENST00000409410.2
ENST00000215069.4
|
CATSPERG
|
catsper channel auxiliary subunit gamma
|
|
chr15_+_59903975
Show fit
|
0.15 |
ENST00000560585.1
ENST00000396065.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr19_+_2841433
Show fit
|
0.15 |
ENST00000334241.4
ENST00000585966.1
ENST00000591539.1
|
ZNF555
|
zinc finger protein 555
|
|
chr5_+_61708582
Show fit
|
0.15 |
ENST00000325324.6
|
IPO11
|
importin 11
|
|
chr10_+_134232503
Show fit
|
0.14 |
ENST00000428814.1
|
RP11-432J24.2
|
RP11-432J24.2
|
|
chr16_+_28770025
Show fit
|
0.14 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9
|
|
chr3_-_42845951
Show fit
|
0.14 |
ENST00000418900.2
ENST00000430190.1
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr17_+_73780852
Show fit
|
0.14 |
ENST00000589666.1
|
UNK
|
unkempt family zinc finger
|
|
chr4_-_170533723
Show fit
|
0.14 |
ENST00000510533.1
ENST00000439128.2
ENST00000511633.1
ENST00000512193.1
ENST00000507142.1
|
NEK1
|
NIMA-related kinase 1
|
|
chr19_+_17326521
Show fit
|
0.14 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr18_+_20494078
Show fit
|
0.14 |
ENST00000579124.1
ENST00000577588.1
ENST00000582354.1
ENST00000581819.1
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr4_+_113218497
Show fit
|
0.14 |
ENST00000458497.1
ENST00000177648.9
ENST00000504176.2
|
ALPK1
|
alpha-kinase 1
|
|
chr4_-_157892498
Show fit
|
0.14 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C
|
|
chr11_+_114270752
Show fit
|
0.14 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7
|