Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
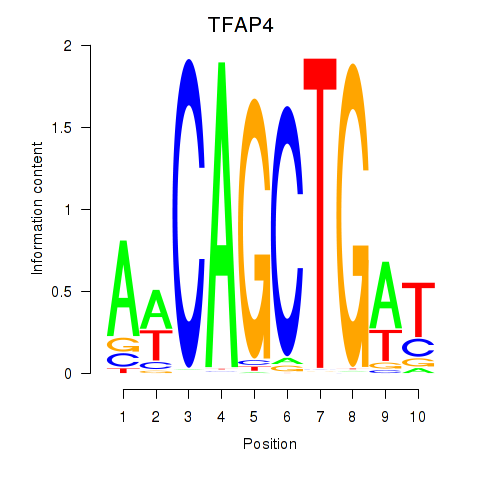
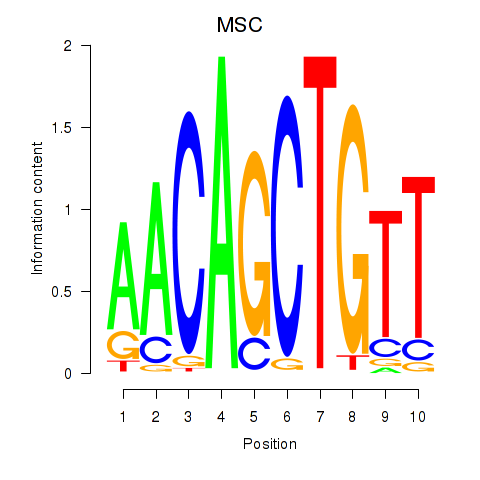
Results for TFAP4_MSC
Z-value: 0.60
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.7 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.8 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg19_v2_chr8_-_72756667_72756736 | -0.77 | 7.6e-02 | Click! |
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | 0.23 | 6.6e-01 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_17791648 | 0.55 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_203256898 | 0.40 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chrX_+_70521584 | 0.33 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr2_-_164592497 | 0.32 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr15_-_40401062 | 0.30 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr8_-_72274467 | 0.26 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr8_-_72274095 | 0.26 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr14_-_81902516 | 0.25 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr1_-_219615984 | 0.25 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr12_-_68845417 | 0.24 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr12_+_26274917 | 0.22 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr3_-_58200398 | 0.22 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr4_+_86699834 | 0.21 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr9_-_33447584 | 0.21 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr14_-_81902791 | 0.20 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr21_+_17791838 | 0.20 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_36508111 | 0.19 |
ENST00000331159.5
ENST00000577233.1 |
SOCS7
|
suppressor of cytokine signaling 7 |
| chr8_-_95961578 | 0.19 |
ENST00000448464.2
ENST00000342697.4 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_+_90272488 | 0.19 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr4_+_108815402 | 0.19 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chrX_-_108868390 | 0.18 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr8_-_72274355 | 0.18 |
ENST00000388741.2
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_-_117021430 | 0.18 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr11_-_85430356 | 0.18 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr3_+_3841108 | 0.17 |
ENST00000319331.3
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chr4_+_154178520 | 0.17 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr11_-_85430204 | 0.17 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_-_153303658 | 0.16 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_+_50679506 | 0.16 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr11_+_33061336 | 0.16 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr4_-_52904425 | 0.16 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr16_-_21289627 | 0.16 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr14_-_71107921 | 0.15 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr17_+_39421591 | 0.15 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr6_+_155470243 | 0.15 |
ENST00000456877.2
ENST00000528391.2 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr16_-_79634595 | 0.15 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr1_-_205391178 | 0.15 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr6_-_39197226 | 0.15 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr9_+_125795788 | 0.15 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr17_+_4981535 | 0.15 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr2_-_42180940 | 0.15 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr3_-_178789220 | 0.14 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr1_+_86889769 | 0.14 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr2_+_233404429 | 0.14 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr10_-_73848764 | 0.14 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr12_+_72233487 | 0.14 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr8_+_87111059 | 0.14 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr19_+_18208603 | 0.14 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr2_-_227050079 | 0.13 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr3_-_49058479 | 0.13 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr8_-_121824374 | 0.13 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr20_-_56195525 | 0.13 |
ENST00000371173.3
ENST00000395822.3 ENST00000340462.4 ENST00000343535.4 |
ZBP1
|
Z-DNA binding protein 1 |
| chr11_-_66084508 | 0.13 |
ENST00000311330.3
|
CD248
|
CD248 molecule, endosialin |
| chr12_+_56473628 | 0.13 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_-_85430088 | 0.13 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_42384343 | 0.12 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr3_+_32148106 | 0.12 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr11_+_124933191 | 0.12 |
ENST00000532000.1
ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr15_-_75748143 | 0.12 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr5_+_102201687 | 0.12 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_-_85430163 | 0.12 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr9_+_131703757 | 0.12 |
ENST00000482796.1
|
RP11-101E3.5
|
RP11-101E3.5 |
| chr17_+_70036164 | 0.12 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr16_+_83932684 | 0.12 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr12_-_54121261 | 0.11 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr17_+_34901353 | 0.11 |
ENST00000593016.1
|
GGNBP2
|
gametogenetin binding protein 2 |
| chr12_-_70093235 | 0.11 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr15_+_49715293 | 0.11 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chrX_-_107019181 | 0.11 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr11_-_111783595 | 0.11 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr6_+_155537771 | 0.10 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr17_+_42925270 | 0.10 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr2_+_54198210 | 0.10 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr1_-_150669604 | 0.10 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr11_-_1785139 | 0.10 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr1_-_9129598 | 0.10 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr15_+_75487984 | 0.10 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr8_+_86351056 | 0.10 |
ENST00000285381.2
|
CA3
|
carbonic anhydrase III, muscle specific |
| chr1_-_2461684 | 0.10 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr2_+_33701707 | 0.10 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_212738676 | 0.10 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr1_+_78354330 | 0.10 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr15_-_52944231 | 0.10 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr1_+_78354175 | 0.09 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr11_-_123185475 | 0.09 |
ENST00000527774.1
ENST00000527533.1 |
RP11-109E10.1
|
RP11-109E10.1 |
| chr10_+_22605374 | 0.09 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr5_+_102201722 | 0.09 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_9129631 | 0.09 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_+_124429006 | 0.09 |
ENST00000522194.1
ENST00000523356.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr12_-_54121212 | 0.09 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr1_+_78354297 | 0.09 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr19_+_32836499 | 0.09 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr10_+_1102303 | 0.09 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr14_-_77495007 | 0.09 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr19_+_7413835 | 0.09 |
ENST00000576789.1
|
CTB-133G6.1
|
CTB-133G6.1 |
| chr2_+_233562015 | 0.09 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chrX_-_101397433 | 0.09 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chr4_+_47487285 | 0.09 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr15_+_75628394 | 0.09 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr14_+_53019993 | 0.09 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr2_-_176867534 | 0.09 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr3_-_178790057 | 0.09 |
ENST00000311417.2
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chrX_-_153210107 | 0.08 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chrX_+_107068959 | 0.08 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr5_+_68463043 | 0.08 |
ENST00000508407.1
ENST00000505500.1 |
CCNB1
|
cyclin B1 |
| chr17_-_39258461 | 0.08 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr5_-_138775177 | 0.08 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_+_138209688 | 0.08 |
ENST00000518381.1
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr1_+_150039877 | 0.08 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr13_-_41635512 | 0.08 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_-_40349106 | 0.08 |
ENST00000545233.1
ENST00000537440.1 ENST00000537223.1 ENST00000541099.1 ENST00000441669.2 ENST00000544981.1 ENST00000316891.5 ENST00000372818.1 |
TRIT1
|
tRNA isopentenyltransferase 1 |
| chrX_-_150067069 | 0.08 |
ENST00000466436.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr6_-_24911195 | 0.08 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr19_-_49176264 | 0.08 |
ENST00000270235.4
ENST00000596844.1 |
NTN5
|
netrin 5 |
| chr6_+_41021027 | 0.08 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr4_+_39408470 | 0.08 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr20_-_40247002 | 0.08 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr16_+_290181 | 0.08 |
ENST00000417499.1
|
ITFG3
|
integrin alpha FG-GAP repeat containing 3 |
| chr9_+_6757634 | 0.08 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr2_+_136343904 | 0.08 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr17_+_67410832 | 0.08 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_14075903 | 0.08 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr15_-_67546963 | 0.08 |
ENST00000561452.1
ENST00000261880.5 |
AAGAB
|
alpha- and gamma-adaptin binding protein |
| chr5_+_95066823 | 0.08 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr10_+_81272287 | 0.08 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr2_+_95537248 | 0.07 |
ENST00000427593.2
|
TEKT4
|
tektin 4 |
| chr5_+_102201509 | 0.07 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_+_89628778 | 0.07 |
ENST00000472354.1
|
RPL13
|
ribosomal protein L13 |
| chr10_+_88428206 | 0.07 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr9_-_35619539 | 0.07 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr19_+_10527449 | 0.07 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chrX_+_53078465 | 0.07 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr4_-_6474173 | 0.07 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr4_+_77941685 | 0.07 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr4_-_108204846 | 0.07 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr4_+_169633310 | 0.07 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_+_1102721 | 0.07 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr1_+_174844645 | 0.07 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr19_+_36249057 | 0.07 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr5_-_173173171 | 0.07 |
ENST00000517733.1
ENST00000518902.1 |
CTB-43E15.3
|
CTB-43E15.3 |
| chr12_-_39299406 | 0.07 |
ENST00000331366.5
|
CPNE8
|
copine VIII |
| chr17_+_44679808 | 0.07 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr2_-_69870747 | 0.07 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr1_+_14075865 | 0.07 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chrX_-_111923145 | 0.07 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr3_-_64211112 | 0.07 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr17_-_39459103 | 0.07 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr20_+_44098385 | 0.07 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr8_+_9413410 | 0.07 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr3_+_159570722 | 0.07 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr9_+_69650263 | 0.07 |
ENST00000322495.3
|
AL445665.1
|
Protein LOC100996643 |
| chr7_-_100171270 | 0.07 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr19_+_46002868 | 0.07 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_-_17981462 | 0.07 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr11_-_1507965 | 0.07 |
ENST00000329957.6
|
MOB2
|
MOB kinase activator 2 |
| chr1_+_185126291 | 0.07 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr5_+_68462944 | 0.07 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr5_+_43603229 | 0.07 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr10_-_73611046 | 0.07 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr14_+_104710541 | 0.07 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr4_+_142558078 | 0.06 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr2_-_202645612 | 0.06 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chrX_+_107069063 | 0.06 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr12_+_104458235 | 0.06 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr15_-_75748115 | 0.06 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr4_+_619347 | 0.06 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr8_+_81398444 | 0.06 |
ENST00000455036.3
ENST00000426744.2 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr10_+_22605304 | 0.06 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr4_+_619386 | 0.06 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr16_-_31105870 | 0.06 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr8_-_110660999 | 0.06 |
ENST00000424158.2
ENST00000533895.1 ENST00000446070.2 ENST00000528331.1 ENST00000526302.1 ENST00000433638.1 ENST00000408908.2 ENST00000524720.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr18_+_19668021 | 0.06 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr20_+_44098346 | 0.06 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr13_+_41635617 | 0.06 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr11_-_14913190 | 0.06 |
ENST00000532378.1
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr1_-_31230650 | 0.06 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr1_-_19638566 | 0.06 |
ENST00000330072.5
ENST00000235835.3 |
AKR7A2
|
aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) |
| chr16_-_4323015 | 0.06 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr12_-_65153175 | 0.06 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr20_-_50418972 | 0.06 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_75628232 | 0.06 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr5_+_102201430 | 0.06 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_95848824 | 0.06 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr2_+_200472779 | 0.06 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr10_-_102090243 | 0.06 |
ENST00000338519.3
ENST00000353274.3 ENST00000318222.3 |
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr7_-_99869799 | 0.06 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr11_+_6411636 | 0.06 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr15_-_55562451 | 0.06 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_107318157 | 0.06 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chrX_+_69509927 | 0.06 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr12_-_4647606 | 0.06 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr1_-_116926718 | 0.06 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chr18_-_74728998 | 0.06 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr17_+_57233087 | 0.06 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr10_+_88428370 | 0.06 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr4_+_159727222 | 0.06 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr10_-_92681033 | 0.06 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr5_+_68462837 | 0.05 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr19_-_46285646 | 0.05 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr17_-_27467418 | 0.05 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:0046680 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.2 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.2 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |