Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
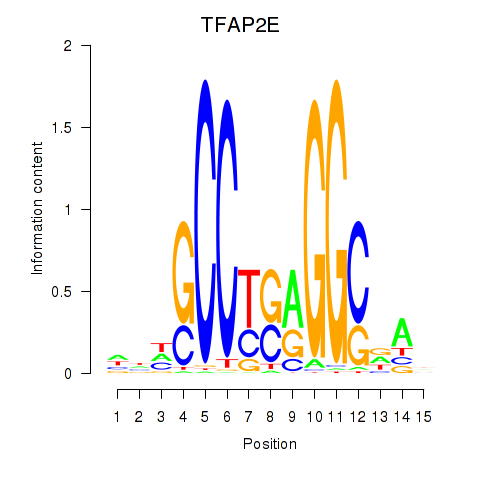
Results for TFAP2E
Z-value: 0.35
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2E | hg19_v2_chr1_+_36038971_36038971 | -0.74 | 9.1e-02 | Click! |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_23810903 | 0.30 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr8_+_22250334 | 0.20 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr20_+_36888551 | 0.17 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr9_-_34662651 | 0.15 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr12_-_15942309 | 0.14 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_-_231175964 | 0.13 |
ENST00000366654.4
|
FAM89A
|
family with sequence similarity 89, member A |
| chr11_-_82612549 | 0.12 |
ENST00000528082.1
ENST00000533126.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_-_35981358 | 0.11 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr3_+_50192833 | 0.11 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr12_-_15942503 | 0.11 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_104068312 | 0.10 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_+_104068562 | 0.10 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr2_-_45236540 | 0.10 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr12_+_53443963 | 0.10 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_+_38893047 | 0.10 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr20_-_30539773 | 0.09 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr6_+_292051 | 0.09 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr12_+_75784850 | 0.09 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr16_-_72698834 | 0.08 |
ENST00000570152.1
ENST00000561611.2 ENST00000570035.1 |
AC004158.2
|
AC004158.2 |
| chr22_+_22676808 | 0.08 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr7_+_100464760 | 0.08 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr17_+_72428218 | 0.08 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr17_+_74070853 | 0.07 |
ENST00000329003.3
|
GALR2
|
galanin receptor 2 |
| chr11_-_82612727 | 0.07 |
ENST00000531128.1
ENST00000535099.1 ENST00000527444.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr10_-_98273668 | 0.07 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr6_+_13272904 | 0.07 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_+_19230433 | 0.07 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr11_-_82612678 | 0.07 |
ENST00000534631.1
ENST00000531801.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_+_3572925 | 0.07 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr2_-_174828892 | 0.07 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chrX_+_152990302 | 0.07 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr3_+_4535025 | 0.07 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_+_54960358 | 0.07 |
ENST00000439657.1
ENST00000376514.2 ENST00000376526.4 ENST00000436479.1 |
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr4_-_82393052 | 0.07 |
ENST00000335927.7
ENST00000504863.1 ENST00000264400.2 |
RASGEF1B
|
RasGEF domain family, member 1B |
| chr19_-_11039188 | 0.06 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr11_+_63974135 | 0.06 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr6_-_88411911 | 0.06 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr11_-_65325430 | 0.06 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr2_+_241375069 | 0.06 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr22_+_38146987 | 0.06 |
ENST00000418339.1
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr3_-_48632593 | 0.06 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr11_-_61062762 | 0.05 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr3_+_105086056 | 0.05 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr19_-_11039261 | 0.05 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr8_+_17780483 | 0.05 |
ENST00000517730.1
ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1
|
pericentriolar material 1 |
| chr19_-_39264072 | 0.05 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr10_-_98480243 | 0.05 |
ENST00000339364.5
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr5_-_179498703 | 0.05 |
ENST00000522208.2
|
RNF130
|
ring finger protein 130 |
| chr11_+_118478313 | 0.05 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr5_-_179719048 | 0.05 |
ENST00000523583.1
ENST00000393360.3 ENST00000455781.1 ENST00000347470.4 ENST00000452135.2 ENST00000343111.6 |
MAPK9
|
mitogen-activated protein kinase 9 |
| chr19_-_54676884 | 0.05 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr19_-_54676846 | 0.04 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr1_+_211433275 | 0.04 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr9_-_21077939 | 0.04 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr17_+_36584662 | 0.04 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr15_+_23810853 | 0.04 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr6_+_42531798 | 0.04 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr8_+_17780346 | 0.04 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr20_-_25371412 | 0.04 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr2_-_177502254 | 0.04 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr1_+_901847 | 0.04 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_-_179499086 | 0.03 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr16_+_69600058 | 0.03 |
ENST00000393742.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr17_+_48423453 | 0.03 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr13_-_76056250 | 0.03 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr12_-_65153175 | 0.03 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr4_+_154387480 | 0.03 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr16_+_69599861 | 0.03 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr19_+_3572758 | 0.03 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr17_+_40932610 | 0.03 |
ENST00000246914.5
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr17_+_37824217 | 0.03 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr16_+_69599899 | 0.03 |
ENST00000567239.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr2_-_24149977 | 0.02 |
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr1_+_17248418 | 0.02 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr10_+_99894380 | 0.02 |
ENST00000370584.3
|
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr19_-_10613862 | 0.02 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_+_38794797 | 0.02 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr14_-_64010046 | 0.02 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr17_+_37824411 | 0.02 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr1_-_201346761 | 0.02 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr7_-_100065686 | 0.02 |
ENST00000423266.1
ENST00000456330.1 |
TSC22D4
|
TSC22 domain family, member 4 |
| chr1_+_233749739 | 0.02 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr1_-_114355083 | 0.02 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr10_-_112678904 | 0.02 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr17_-_42188598 | 0.02 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chrX_+_131157322 | 0.02 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr3_+_155860751 | 0.02 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_74741569 | 0.01 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr20_+_55205825 | 0.01 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr10_+_99894399 | 0.01 |
ENST00000298999.3
ENST00000314594.5 |
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr19_+_4969116 | 0.01 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr2_+_38893208 | 0.01 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr13_-_27334879 | 0.01 |
ENST00000405846.3
|
GPR12
|
G protein-coupled receptor 12 |
| chrX_+_131157290 | 0.01 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr19_-_46389359 | 0.01 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr20_+_58508817 | 0.01 |
ENST00000358293.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr5_-_179499108 | 0.01 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr7_-_129251459 | 0.01 |
ENST00000608694.1
|
RP11-448A19.1
|
RP11-448A19.1 |
| chrX_+_49028265 | 0.01 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr10_-_108924284 | 0.01 |
ENST00000344440.6
ENST00000263054.6 |
SORCS1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr11_-_35441524 | 0.01 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_140624499 | 0.01 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr11_-_66056596 | 0.01 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chrX_-_118826784 | 0.01 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr6_+_37787262 | 0.00 |
ENST00000287218.4
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr12_+_53773944 | 0.00 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr17_-_31620006 | 0.00 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr4_-_101439242 | 0.00 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr11_-_6633799 | 0.00 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr11_+_82612740 | 0.00 |
ENST00000524921.1
ENST00000528759.1 ENST00000525361.1 ENST00000430323.2 ENST00000533655.1 ENST00000532764.1 ENST00000532589.1 ENST00000525388.1 |
C11orf82
|
chromosome 11 open reading frame 82 |
| chr3_-_55521323 | 0.00 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chrX_+_51636629 | 0.00 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr17_+_29158962 | 0.00 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr11_-_71753188 | 0.00 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr17_-_42992856 | 0.00 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.0 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |