Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
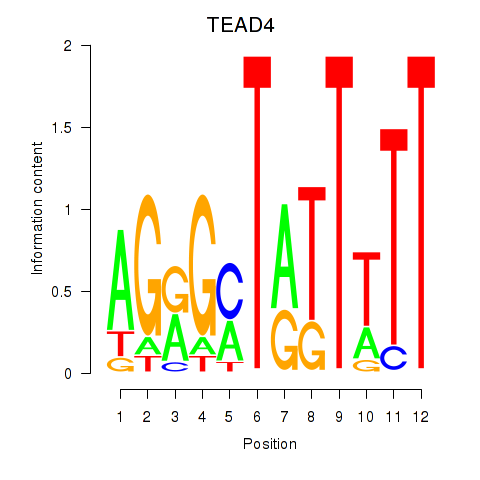
Results for TEAD4
Z-value: 0.52
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.4 | TEA domain transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg19_v2_chr12_+_3068466_3068496 | 0.22 | 6.8e-01 | Click! |
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_110723134 | 0.25 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr10_+_90484301 | 0.25 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr10_-_4285923 | 0.24 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr4_-_110723335 | 0.22 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_110723194 | 0.21 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr12_+_28605426 | 0.20 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr4_+_75174180 | 0.19 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr19_+_57631411 | 0.18 |
ENST00000254181.4
ENST00000600940.1 |
USP29
|
ubiquitin specific peptidase 29 |
| chr11_+_1295809 | 0.18 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr10_-_4285835 | 0.16 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr4_+_186317133 | 0.15 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr13_-_38172863 | 0.15 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr6_+_122720681 | 0.15 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr9_+_67968793 | 0.13 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr15_-_83224682 | 0.13 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr9_-_15472730 | 0.12 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr18_-_25616519 | 0.12 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chrX_+_49832231 | 0.11 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr11_-_59633951 | 0.11 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr1_-_207119738 | 0.11 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr15_+_81475047 | 0.11 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr13_+_20268547 | 0.11 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr2_+_108905325 | 0.11 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_+_186692745 | 0.11 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr9_+_71944241 | 0.11 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr2_+_196440692 | 0.10 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr22_+_24115000 | 0.10 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chrX_+_22056165 | 0.10 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_-_159080806 | 0.10 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr7_+_18548924 | 0.09 |
ENST00000524023.1
|
HDAC9
|
histone deacetylase 9 |
| chr1_+_206623784 | 0.09 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_121973974 | 0.09 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr21_+_17909594 | 0.09 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_56367101 | 0.09 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr12_-_49333446 | 0.09 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr18_+_61254221 | 0.08 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr15_+_59908633 | 0.08 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr10_+_63808970 | 0.08 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr12_+_42725554 | 0.08 |
ENST00000546750.1
ENST00000547847.1 |
PPHLN1
|
periphilin 1 |
| chr12_+_123011776 | 0.07 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr1_+_164528616 | 0.07 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_241695670 | 0.07 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_145425573 | 0.07 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_-_10308125 | 0.07 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr3_-_164875850 | 0.07 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr6_+_46761118 | 0.07 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr4_-_186317034 | 0.07 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr6_+_151358048 | 0.07 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr5_+_173472607 | 0.07 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr5_+_136070614 | 0.07 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr7_+_23719749 | 0.06 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr9_-_119162885 | 0.06 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr4_-_155533787 | 0.06 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr14_+_76071805 | 0.06 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr1_+_241695424 | 0.06 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_+_52074502 | 0.06 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr2_-_42588289 | 0.06 |
ENST00000468711.1
ENST00000463055.1 |
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr21_+_34804479 | 0.06 |
ENST00000421802.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr7_-_14029283 | 0.06 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr20_+_33814457 | 0.06 |
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted) |
| chr2_-_48468122 | 0.06 |
ENST00000447571.1
|
AC079807.4
|
AC079807.4 |
| chr11_+_86502085 | 0.06 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr1_-_79472365 | 0.06 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr1_-_154928562 | 0.06 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr11_-_92930556 | 0.06 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr16_-_46655538 | 0.06 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr15_+_59730348 | 0.06 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr1_+_171283331 | 0.06 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr11_-_79151695 | 0.06 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr2_+_145425534 | 0.06 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr4_+_96012585 | 0.06 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr21_+_17443521 | 0.06 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_+_2029019 | 0.05 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_100566492 | 0.05 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chrX_+_37639264 | 0.05 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_+_178310581 | 0.05 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr6_-_109330702 | 0.05 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr5_-_111093167 | 0.05 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr15_+_80733570 | 0.05 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_164528866 | 0.05 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_174670143 | 0.05 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_-_188419078 | 0.05 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr15_-_38852251 | 0.05 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr8_+_107460147 | 0.05 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr2_-_161056802 | 0.05 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr9_-_104145795 | 0.05 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr4_-_89152474 | 0.05 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr6_-_27835357 | 0.04 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr2_-_196933536 | 0.04 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr3_-_25915189 | 0.04 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr5_-_138775177 | 0.04 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr12_-_2113583 | 0.04 |
ENST00000397173.4
ENST00000280665.6 |
DCP1B
|
decapping mRNA 1B |
| chr12_+_48876275 | 0.04 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr10_+_15074190 | 0.04 |
ENST00000428897.1
ENST00000413672.1 |
OLAH
|
oleoyl-ACP hydrolase |
| chr5_+_140261703 | 0.04 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr3_-_112356944 | 0.04 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr8_+_107282368 | 0.04 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr9_-_96215822 | 0.04 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr3_+_123813509 | 0.04 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr3_-_11685345 | 0.04 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_-_159466042 | 0.04 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr18_+_61254570 | 0.04 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr12_-_54652060 | 0.04 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr12_+_10460417 | 0.04 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr8_+_92114060 | 0.04 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr7_-_16872932 | 0.04 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr2_-_161056762 | 0.04 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr6_-_56507586 | 0.04 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr2_-_211341411 | 0.04 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr3_+_38307293 | 0.04 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr7_-_111428957 | 0.04 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr10_-_119805558 | 0.04 |
ENST00000369199.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr6_-_152489484 | 0.04 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_164529004 | 0.04 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_-_47983519 | 0.04 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr22_+_46449674 | 0.04 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr3_-_137834436 | 0.04 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr4_-_149365827 | 0.04 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr17_+_79369249 | 0.03 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr5_-_124081008 | 0.03 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr4_+_40337340 | 0.03 |
ENST00000310169.2
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
| chr1_+_107683436 | 0.03 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr8_-_30002179 | 0.03 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr11_-_6440624 | 0.03 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_63376013 | 0.03 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr15_+_100106155 | 0.03 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr10_-_90712520 | 0.03 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr10_+_123923205 | 0.03 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr5_+_138609782 | 0.03 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr9_+_131902283 | 0.03 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr9_+_42671887 | 0.03 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr5_-_138862326 | 0.03 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr12_-_58220078 | 0.03 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr6_-_41039567 | 0.03 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr16_+_4364762 | 0.03 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr10_-_52008313 | 0.03 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr1_+_236557569 | 0.03 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr2_-_99279928 | 0.03 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_+_239047337 | 0.03 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chr1_+_114473350 | 0.03 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr12_-_111806892 | 0.03 |
ENST00000547710.1
ENST00000549321.1 ENST00000361483.3 ENST00000392658.5 |
FAM109A
|
family with sequence similarity 109, member A |
| chr11_+_94439591 | 0.03 |
ENST00000299004.9
|
AMOTL1
|
angiomotin like 1 |
| chr9_+_90112117 | 0.03 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr4_+_141264597 | 0.03 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr11_+_125496400 | 0.03 |
ENST00000524737.1
|
CHEK1
|
checkpoint kinase 1 |
| chr12_-_49393092 | 0.03 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr12_+_97306295 | 0.03 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr12_+_109554386 | 0.03 |
ENST00000338432.7
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr17_-_39140549 | 0.03 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr1_+_32716840 | 0.03 |
ENST00000336890.5
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr1_-_151798546 | 0.03 |
ENST00000356728.6
|
RORC
|
RAR-related orphan receptor C |
| chr12_+_56511943 | 0.03 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr19_+_37825526 | 0.03 |
ENST00000592768.1
ENST00000591417.1 ENST00000585623.1 ENST00000592168.1 ENST00000591391.1 ENST00000392153.3 ENST00000589392.1 ENST00000324411.4 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr12_+_70574088 | 0.02 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr12_+_32260085 | 0.02 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr21_-_39870339 | 0.02 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr6_-_134639180 | 0.02 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_42588338 | 0.02 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr11_+_17316870 | 0.02 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr7_-_14028488 | 0.02 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr4_-_100485183 | 0.02 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr8_+_107282389 | 0.02 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr15_+_100106244 | 0.02 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr4_+_96012614 | 0.02 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr18_+_6834472 | 0.02 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr7_+_123488124 | 0.02 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr16_-_46797149 | 0.02 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr2_+_238767517 | 0.02 |
ENST00000404910.2
|
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1 |
| chr4_-_186732048 | 0.02 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_164528437 | 0.02 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr2_-_237150258 | 0.02 |
ENST00000330842.6
|
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr7_-_45956856 | 0.02 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr8_+_66619277 | 0.02 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr4_+_160203650 | 0.02 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_87563557 | 0.02 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr2_-_231989808 | 0.02 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr10_-_75410771 | 0.02 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr19_+_37837185 | 0.02 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr4_+_141178440 | 0.02 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr2_+_121493717 | 0.02 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr10_+_93683519 | 0.02 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr2_+_197577841 | 0.02 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr8_+_42128861 | 0.02 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr7_+_152456829 | 0.02 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chrX_-_15402498 | 0.02 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr3_-_18466026 | 0.02 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr7_+_37960163 | 0.02 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr20_+_47538357 | 0.02 |
ENST00000371917.4
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr11_+_125496619 | 0.02 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr8_+_24151620 | 0.02 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_-_91572278 | 0.02 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr6_-_152958521 | 0.02 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_-_128568721 | 0.02 |
ENST00000322313.4
ENST00000393006.1 ENST00000409658.3 ENST00000436787.1 |
WDR33
|
WD repeat domain 33 |
| chr4_+_41614909 | 0.02 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr15_-_72563585 | 0.02 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr4_-_89978299 | 0.02 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr2_-_188419200 | 0.02 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr10_-_62149433 | 0.01 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_+_68286516 | 0.01 |
ENST00000471583.1
ENST00000487270.1 ENST00000488612.1 |
RAD51B
|
RAD51 paralog B |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:2000809 | regulation of postsynaptic density protein 95 clustering(GO:1902897) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |