Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
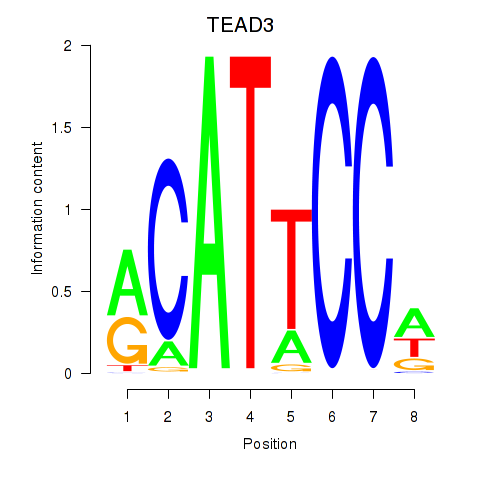
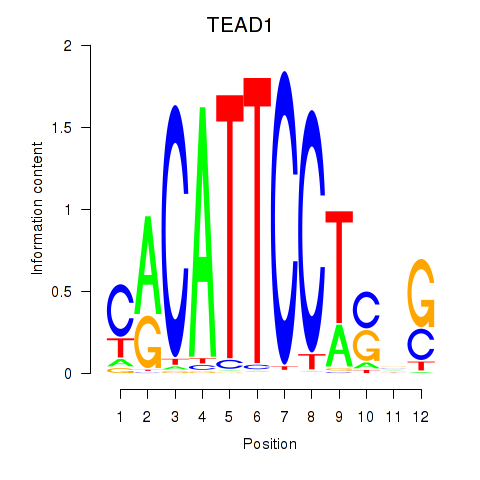
Results for TEAD3_TEAD1
Z-value: 0.38
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEA domain transcription factor 3 |
|
TEAD1
|
ENSG00000187079.10 | TEA domain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg19_v2_chr11_+_12696102_12696164 | -0.23 | 6.6e-01 | Click! |
| TEAD3 | hg19_v2_chr6_-_35464727_35464738 | -0.04 | 9.4e-01 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_69590486 | 0.35 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr4_-_186732892 | 0.24 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_-_90712520 | 0.23 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr4_+_165675197 | 0.22 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr17_-_65242063 | 0.21 |
ENST00000581159.1
|
HELZ
|
helicase with zinc finger |
| chr13_+_113030658 | 0.20 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr17_-_39677971 | 0.15 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr17_-_58603482 | 0.14 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr11_-_85430356 | 0.13 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr17_-_39459103 | 0.13 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr9_-_34381536 | 0.13 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr10_-_75351088 | 0.13 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr3_-_52869205 | 0.13 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr2_+_208104497 | 0.12 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr6_+_116850174 | 0.12 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr1_+_150229554 | 0.11 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr14_+_62331592 | 0.11 |
ENST00000554436.1
|
CTD-2277K2.1
|
CTD-2277K2.1 |
| chr12_-_127630534 | 0.10 |
ENST00000535022.1
|
RP11-575F12.2
|
RP11-575F12.2 |
| chr2_+_228029281 | 0.10 |
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr12_+_120502441 | 0.10 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr16_+_30907927 | 0.10 |
ENST00000279804.2
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chr9_+_131684027 | 0.10 |
ENST00000426694.1
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_+_29823427 | 0.09 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr6_-_132272504 | 0.09 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr14_+_76044934 | 0.09 |
ENST00000238667.4
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr7_-_2349579 | 0.09 |
ENST00000435060.1
|
SNX8
|
sorting nexin 8 |
| chr22_+_36784632 | 0.09 |
ENST00000424761.1
|
RP4-633O19__A.1
|
RP4-633O19__A.1 |
| chr16_+_47496023 | 0.09 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr5_-_39424961 | 0.09 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_9944492 | 0.08 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr19_-_33182616 | 0.08 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr8_-_102803163 | 0.08 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr12_-_8815299 | 0.08 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr9_+_72658490 | 0.08 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr20_-_45530365 | 0.08 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr9_-_127905736 | 0.08 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr4_+_165675269 | 0.08 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr8_+_70378852 | 0.08 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr2_+_145780767 | 0.08 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr7_+_102715315 | 0.08 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr5_+_95998714 | 0.08 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr6_-_10435032 | 0.07 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr2_-_192711968 | 0.07 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr1_-_161279749 | 0.07 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chrX_-_108868390 | 0.07 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr15_+_96869165 | 0.07 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_169769787 | 0.07 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr14_+_21566980 | 0.07 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr6_+_158402860 | 0.07 |
ENST00000367122.2
ENST00000367121.3 ENST00000355585.4 ENST00000367113.4 |
SYNJ2
|
synaptojanin 2 |
| chr7_-_102715172 | 0.07 |
ENST00000456695.1
ENST00000455112.2 ENST00000440067.1 |
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr12_+_27677085 | 0.06 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr10_+_99344071 | 0.06 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr4_-_40632140 | 0.06 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr4_-_186733363 | 0.06 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_44037546 | 0.06 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr1_+_12538594 | 0.06 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr17_-_39123144 | 0.06 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr14_+_65878650 | 0.06 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr2_-_75788428 | 0.06 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_+_208104351 | 0.06 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr12_-_7245152 | 0.06 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr19_-_49567124 | 0.06 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr12_-_56359802 | 0.06 |
ENST00000548803.1
ENST00000449260.2 ENST00000550447.1 ENST00000547137.1 ENST00000546543.1 ENST00000550464.1 |
PMEL
|
premelanosome protein |
| chr5_-_150138246 | 0.06 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chr11_-_62521614 | 0.06 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr12_-_118490403 | 0.06 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr7_-_100881041 | 0.06 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr10_+_81891416 | 0.06 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr19_-_51327034 | 0.06 |
ENST00000301420.2
ENST00000448701.2 |
KLK1
|
kallikrein 1 |
| chr6_+_143999185 | 0.06 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr8_-_101321584 | 0.06 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr16_+_30386098 | 0.06 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr10_+_112404132 | 0.05 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr8_-_108510224 | 0.05 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr1_+_62417957 | 0.05 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr9_+_95736758 | 0.05 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr11_+_66824303 | 0.05 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr4_+_154178520 | 0.05 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr17_-_8079632 | 0.05 |
ENST00000431792.2
|
TMEM107
|
transmembrane protein 107 |
| chr14_+_22471345 | 0.05 |
ENST00000390446.3
|
TRAV18
|
T cell receptor alpha variable 18 |
| chr12_-_123565834 | 0.05 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr7_+_100464760 | 0.05 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr5_-_150138061 | 0.05 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr10_-_52008313 | 0.05 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr9_-_130700080 | 0.05 |
ENST00000373110.4
|
DPM2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chrX_-_6146876 | 0.05 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr2_-_75788038 | 0.05 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr4_-_57301748 | 0.05 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr2_+_189157536 | 0.05 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr9_+_71357171 | 0.05 |
ENST00000440050.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr11_-_8892900 | 0.05 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr17_-_42767092 | 0.05 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr1_+_154401791 | 0.05 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr3_-_52868931 | 0.05 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr6_+_46761118 | 0.05 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr2_-_164592497 | 0.05 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr12_-_63328817 | 0.05 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr6_+_35996859 | 0.05 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr15_-_32747103 | 0.05 |
ENST00000562377.1
|
GOLGA8O
|
golgin A8 family, member O |
| chr2_+_170550944 | 0.05 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr4_+_110834033 | 0.04 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr15_+_41549105 | 0.04 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr20_+_35169885 | 0.04 |
ENST00000279022.2
ENST00000346786.2 |
MYL9
|
myosin, light chain 9, regulatory |
| chr5_+_95998746 | 0.04 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr2_+_189157498 | 0.04 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_-_186732048 | 0.04 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_116165754 | 0.04 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr1_+_107682629 | 0.04 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr11_-_111781554 | 0.04 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr14_-_23623577 | 0.04 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr17_+_20059358 | 0.04 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_-_85780853 | 0.04 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr4_-_152149033 | 0.04 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr14_-_23395623 | 0.04 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr15_+_44719996 | 0.04 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_+_27062860 | 0.04 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr8_+_42873548 | 0.04 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr15_-_77197620 | 0.04 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr6_-_152489484 | 0.04 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr11_-_85430204 | 0.04 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr12_-_8815404 | 0.04 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr15_-_48937982 | 0.04 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr8_+_70379072 | 0.04 |
ENST00000529134.1
|
SULF1
|
sulfatase 1 |
| chr2_-_75788424 | 0.04 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr16_-_30125177 | 0.04 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr17_-_8021710 | 0.04 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr4_-_70725856 | 0.04 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr5_-_54988559 | 0.04 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr11_+_46316677 | 0.04 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr11_-_14379997 | 0.04 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr18_-_53303123 | 0.04 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr1_+_152730499 | 0.04 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr3_-_119379719 | 0.04 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr8_-_29592736 | 0.04 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr7_+_134464414 | 0.04 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr9_+_131684562 | 0.04 |
ENST00000421063.2
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr11_-_119249805 | 0.04 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_+_109271481 | 0.04 |
ENST00000542845.1
ENST00000393314.2 |
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_-_83278322 | 0.04 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_-_218770168 | 0.04 |
ENST00000413554.1
|
TNS1
|
tensin 1 |
| chr12_-_104443890 | 0.04 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr10_-_17659234 | 0.04 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr5_+_36608280 | 0.04 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr15_-_33360342 | 0.04 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr8_-_94753184 | 0.04 |
ENST00000520560.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr1_+_244227632 | 0.04 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr3_-_49058479 | 0.04 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr16_+_70207686 | 0.04 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr11_-_111782484 | 0.04 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr19_+_45409011 | 0.04 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chrX_+_37865804 | 0.04 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr1_+_18807424 | 0.04 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr2_-_161264385 | 0.04 |
ENST00000409972.1
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr16_-_4164027 | 0.04 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr12_-_25150409 | 0.03 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr3_+_25469724 | 0.03 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr11_+_1856034 | 0.03 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr16_-_57570450 | 0.03 |
ENST00000258214.2
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr8_+_97597148 | 0.03 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr1_-_33338076 | 0.03 |
ENST00000496770.1
|
FNDC5
|
fibronectin type III domain containing 5 |
| chr3_-_149375783 | 0.03 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr12_-_58240470 | 0.03 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr5_+_42423872 | 0.03 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr5_+_71475449 | 0.03 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr18_+_32173276 | 0.03 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr11_-_64013288 | 0.03 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr16_-_74455290 | 0.03 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr6_+_108881012 | 0.03 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr2_-_153573965 | 0.03 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_-_185729602 | 0.03 |
ENST00000437665.3
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr10_+_97759848 | 0.03 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr3_+_169940153 | 0.03 |
ENST00000295797.4
|
PRKCI
|
protein kinase C, iota |
| chr1_+_28764653 | 0.03 |
ENST00000373836.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr5_-_146781153 | 0.03 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_+_10312604 | 0.03 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr8_-_101348408 | 0.03 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr10_+_99344104 | 0.03 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr19_-_51472031 | 0.03 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr3_+_25469802 | 0.03 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr9_+_125795788 | 0.03 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr5_+_95998673 | 0.03 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr11_+_1855645 | 0.03 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr7_+_116165038 | 0.03 |
ENST00000393470.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr17_-_70053866 | 0.03 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr11_+_46299199 | 0.03 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr5_+_49962495 | 0.03 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_63921351 | 0.03 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr7_-_25219897 | 0.03 |
ENST00000283905.3
ENST00000409280.1 ENST00000415598.1 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr17_-_57232596 | 0.03 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_-_8815215 | 0.03 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr1_-_95391315 | 0.03 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr12_+_116985896 | 0.03 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr15_+_63334831 | 0.03 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr14_+_32546274 | 0.03 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr9_+_2015335 | 0.03 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_77495007 | 0.03 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr11_+_94439591 | 0.03 |
ENST00000299004.9
|
AMOTL1
|
angiomotin like 1 |
| chr3_-_196159268 | 0.03 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr18_-_25616519 | 0.03 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr2_+_166430619 | 0.03 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.0 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.0 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |