Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
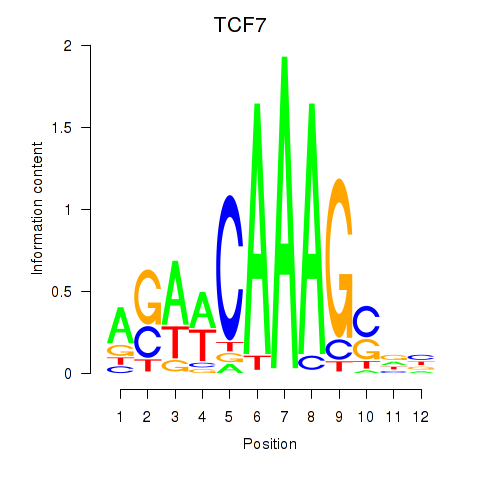
Results for TCF7
Z-value: 0.48
Transcription factors associated with TCF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7
|
ENSG00000081059.15 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7 | hg19_v2_chr5_+_133450365_133450444 | -0.70 | 1.2e-01 | Click! |
Activity profile of TCF7 motif
Sorted Z-values of TCF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_21654714 | 0.42 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr15_+_63414760 | 0.36 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr1_-_153013588 | 0.28 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr1_-_153029980 | 0.26 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr17_-_57229155 | 0.25 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_-_111383064 | 0.23 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr1_-_153113927 | 0.23 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr12_+_100594557 | 0.23 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr2_-_165697717 | 0.19 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr5_+_32531893 | 0.19 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr1_+_200993071 | 0.19 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr21_+_39628780 | 0.18 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_+_111846741 | 0.17 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr7_+_89975979 | 0.17 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr15_-_35280426 | 0.17 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr20_+_5987890 | 0.15 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_-_103266219 | 0.15 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr5_+_136070614 | 0.15 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr17_-_46623441 | 0.14 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr2_+_24150180 | 0.14 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr14_+_61201445 | 0.14 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr12_+_104359576 | 0.13 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr6_+_138188551 | 0.13 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr17_+_34848049 | 0.13 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr7_+_112120908 | 0.12 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr10_-_25241499 | 0.12 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr3_-_197024394 | 0.12 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr7_+_111846643 | 0.12 |
ENST00000361822.3
|
ZNF277
|
zinc finger protein 277 |
| chr17_+_76037081 | 0.12 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr17_+_70026795 | 0.11 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr20_+_45947246 | 0.11 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr1_+_200860122 | 0.11 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr22_+_29168652 | 0.11 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr3_-_120068143 | 0.11 |
ENST00000295628.3
|
LRRC58
|
leucine rich repeat containing 58 |
| chr8_+_97773457 | 0.11 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr12_+_69080734 | 0.11 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr20_+_35504522 | 0.11 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr9_+_91003271 | 0.11 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr6_+_88054530 | 0.11 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr11_-_76155700 | 0.11 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_-_39580775 | 0.10 |
ENST00000225550.3
|
KRT37
|
keratin 37 |
| chr13_+_29233218 | 0.10 |
ENST00000380842.4
|
POMP
|
proteasome maturation protein |
| chr2_-_36825281 | 0.10 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr8_-_124665190 | 0.10 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr12_+_69979446 | 0.09 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr12_-_49582593 | 0.09 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_+_32655048 | 0.09 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_-_16936340 | 0.09 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr5_+_34757309 | 0.09 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr4_+_95128748 | 0.09 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_64281906 | 0.09 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr15_+_30918879 | 0.09 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr19_+_21579908 | 0.09 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr10_+_5090940 | 0.09 |
ENST00000602997.1
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr7_+_151791037 | 0.09 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_+_46926326 | 0.08 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr19_-_44008863 | 0.08 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr7_+_151791074 | 0.08 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_+_220309379 | 0.08 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr8_+_27629459 | 0.08 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_-_90996459 | 0.08 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr13_+_45694583 | 0.08 |
ENST00000340473.6
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa |
| chr13_+_102104952 | 0.08 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_-_42358742 | 0.08 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr12_+_8849773 | 0.08 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr17_-_18266818 | 0.08 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr10_+_115999089 | 0.08 |
ENST00000392982.3
|
VWA2
|
von Willebrand factor A domain containing 2 |
| chr8_+_95653840 | 0.08 |
ENST00000520385.1
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr5_+_135170331 | 0.07 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr12_+_104359614 | 0.07 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr3_+_111718173 | 0.07 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr16_-_29937476 | 0.07 |
ENST00000561540.1
|
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr11_-_33774944 | 0.07 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr1_-_211752073 | 0.07 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr1_-_151345159 | 0.07 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr8_+_55471630 | 0.07 |
ENST00000522001.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr12_-_49581152 | 0.07 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr8_+_56792355 | 0.07 |
ENST00000519728.1
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr14_+_59951161 | 0.07 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr1_-_53793584 | 0.07 |
ENST00000354412.3
ENST00000347547.2 ENST00000306052.6 |
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr11_-_85430088 | 0.07 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr8_+_73921085 | 0.07 |
ENST00000276603.5
ENST00000276602.6 ENST00000518874.1 |
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr11_+_100862811 | 0.07 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr1_+_20208870 | 0.07 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr22_-_42526802 | 0.07 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr4_+_71859156 | 0.07 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr6_-_90062543 | 0.07 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr7_+_151791095 | 0.07 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr22_+_24105394 | 0.07 |
ENST00000305199.5
ENST00000382821.3 |
C22orf15
|
chromosome 22 open reading frame 15 |
| chr9_-_130639997 | 0.07 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr8_+_56792377 | 0.07 |
ENST00000520220.2
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr1_+_75198871 | 0.07 |
ENST00000479111.1
|
TYW3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr4_-_7873713 | 0.07 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr1_+_75198828 | 0.06 |
ENST00000457880.2
ENST00000370867.3 ENST00000421739.2 |
TYW3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr9_+_125137565 | 0.06 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr21_+_30672433 | 0.06 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr9_+_109685630 | 0.06 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr8_-_2585929 | 0.06 |
ENST00000519393.1
ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1
|
RP11-134O21.1 |
| chr12_+_1099675 | 0.06 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr3_-_169487617 | 0.06 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr1_+_15479021 | 0.06 |
ENST00000428417.1
|
TMEM51
|
transmembrane protein 51 |
| chr6_+_43968306 | 0.06 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr7_-_36406750 | 0.06 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr2_+_26568965 | 0.06 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr6_-_10435032 | 0.06 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr11_+_65405556 | 0.06 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr12_-_3982511 | 0.06 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr11_-_85430204 | 0.06 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr6_+_64282447 | 0.06 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr15_-_48470544 | 0.06 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr12_+_112204691 | 0.06 |
ENST00000416293.3
ENST00000261733.2 |
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr2_-_238499337 | 0.06 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr1_+_15479054 | 0.06 |
ENST00000376014.3
ENST00000451326.2 |
TMEM51
|
transmembrane protein 51 |
| chr1_+_59762642 | 0.06 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr4_-_71705027 | 0.06 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr22_+_31742875 | 0.06 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr6_-_112408661 | 0.06 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr6_+_52285046 | 0.06 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr5_-_79287060 | 0.06 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr4_+_95128996 | 0.06 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_80548667 | 0.05 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_+_32655110 | 0.05 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_35704804 | 0.05 |
ENST00000373869.3
|
ARMC12
|
armadillo repeat containing 12 |
| chr15_+_36871983 | 0.05 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr13_+_102104980 | 0.05 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr19_+_10765614 | 0.05 |
ENST00000589283.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr10_-_14614311 | 0.05 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr4_-_71705060 | 0.05 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_+_177134201 | 0.05 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr6_+_35704855 | 0.05 |
ENST00000288065.2
ENST00000373866.3 |
ARMC12
|
armadillo repeat containing 12 |
| chr1_-_62785054 | 0.05 |
ENST00000371153.4
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr8_-_101962777 | 0.05 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr8_-_8751068 | 0.05 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr8_-_42752418 | 0.05 |
ENST00000524954.1
|
RNF170
|
ring finger protein 170 |
| chr17_+_31255023 | 0.05 |
ENST00000395149.2
ENST00000261713.4 |
TMEM98
|
transmembrane protein 98 |
| chr18_+_32455201 | 0.05 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr12_-_14924056 | 0.05 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr8_-_124279627 | 0.05 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr3_+_118892411 | 0.05 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr3_+_111717511 | 0.05 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr12_-_67197760 | 0.05 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr11_-_125365435 | 0.05 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_+_68870721 | 0.05 |
ENST00000303786.3
|
PROKR1
|
prokineticin receptor 1 |
| chr10_-_14613968 | 0.05 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_+_93118813 | 0.05 |
ENST00000556418.1
|
RIN3
|
Ras and Rab interactor 3 |
| chr4_-_106629796 | 0.05 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr19_+_50084561 | 0.05 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr2_-_29093132 | 0.05 |
ENST00000306108.5
|
TRMT61B
|
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr2_-_151344172 | 0.04 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr12_+_104697504 | 0.04 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr1_+_169079823 | 0.04 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr16_-_46864955 | 0.04 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_+_12598563 | 0.04 |
ENST00000411987.1
ENST00000448482.1 |
MKRN2
|
makorin ring finger protein 2 |
| chr16_-_46865047 | 0.04 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr18_+_59992514 | 0.04 |
ENST00000269485.7
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr7_+_150413645 | 0.04 |
ENST00000307194.5
|
GIMAP1
|
GTPase, IMAP family member 1 |
| chr12_+_27677085 | 0.04 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr12_+_104359641 | 0.04 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr4_-_140477928 | 0.04 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr4_+_89300158 | 0.04 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr8_+_38244638 | 0.04 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_95129061 | 0.04 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_-_110933611 | 0.04 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr14_+_100070869 | 0.04 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr19_+_10765699 | 0.04 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr3_+_12598509 | 0.04 |
ENST00000170447.7
|
MKRN2
|
makorin ring finger protein 2 |
| chr5_-_157002749 | 0.04 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr7_+_56119323 | 0.04 |
ENST00000275603.4
ENST00000335503.3 ENST00000540286.1 |
CCT6A
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr11_-_6677018 | 0.04 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr17_+_48611853 | 0.04 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr2_-_238499303 | 0.04 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr14_-_36645674 | 0.04 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr10_+_17271266 | 0.04 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr2_+_46926048 | 0.04 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chrX_-_128977364 | 0.04 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr4_-_74486347 | 0.04 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_+_133879193 | 0.04 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr2_+_181845298 | 0.04 |
ENST00000410062.4
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_-_32801825 | 0.04 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr13_+_102142296 | 0.04 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_58329819 | 0.04 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr1_-_53793725 | 0.03 |
ENST00000371454.2
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_-_33416231 | 0.03 |
ENST00000584655.1
ENST00000447669.2 ENST00000315249.7 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr8_-_37411648 | 0.03 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr12_+_58937907 | 0.03 |
ENST00000546977.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr2_+_201981663 | 0.03 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr7_-_87505658 | 0.03 |
ENST00000341119.5
|
SLC25A40
|
solute carrier family 25, member 40 |
| chr1_-_119869846 | 0.03 |
ENST00000457719.1
|
RP11-418J17.3
|
RP11-418J17.3 |
| chr9_-_21368075 | 0.03 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr5_+_150040403 | 0.03 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr3_-_186285077 | 0.03 |
ENST00000338733.5
|
TBCCD1
|
TBCC domain containing 1 |
| chr2_+_11679963 | 0.03 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr4_+_69917078 | 0.03 |
ENST00000502942.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_-_161695042 | 0.03 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr15_+_74466012 | 0.03 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr6_+_160183492 | 0.03 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr3_+_111718036 | 0.03 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr10_-_14614095 | 0.03 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_-_173991434 | 0.03 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr9_+_103189536 | 0.03 |
ENST00000374885.1
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0070668 | regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0090345 | alkaloid catabolic process(GO:0009822) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.2 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |