Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
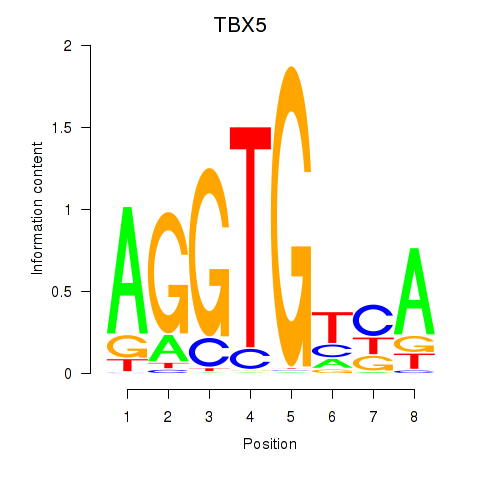
Results for TBX5
Z-value: 0.31
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.15 | T-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg19_v2_chr12_-_114843889_114843921 | 0.28 | 5.9e-01 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_2205352 | 0.20 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr17_-_74533734 | 0.15 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr20_-_4055812 | 0.14 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr11_+_7559485 | 0.14 |
ENST00000527790.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_8579030 | 0.13 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr5_-_74348371 | 0.13 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2 |
| chr17_+_37821593 | 0.13 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr5_-_176836577 | 0.11 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr22_-_36013368 | 0.11 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr14_+_104177607 | 0.11 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chr11_-_45928830 | 0.10 |
ENST00000449465.1
|
C11orf94
|
chromosome 11 open reading frame 94 |
| chr19_+_46531127 | 0.10 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr19_+_55987998 | 0.10 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr1_+_8378140 | 0.10 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr2_+_95963052 | 0.10 |
ENST00000295225.5
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin |
| chr19_-_41934635 | 0.09 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr15_-_74043816 | 0.08 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr2_-_72374948 | 0.08 |
ENST00000546307.1
ENST00000474509.1 |
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr1_-_12679171 | 0.08 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr2_-_241835561 | 0.08 |
ENST00000388934.4
|
C2orf54
|
chromosome 2 open reading frame 54 |
| chr16_-_3086927 | 0.08 |
ENST00000572449.1
|
CCDC64B
|
coiled-coil domain containing 64B |
| chr11_-_115127611 | 0.08 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr19_-_10420459 | 0.08 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr16_-_69385681 | 0.08 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr22_-_20231207 | 0.08 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr6_-_13487825 | 0.08 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr16_+_57139933 | 0.08 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr12_+_57854274 | 0.08 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr4_-_177116772 | 0.08 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr11_-_74660065 | 0.08 |
ENST00000525407.1
ENST00000528219.1 ENST00000531852.1 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr16_-_74808710 | 0.08 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr18_-_59561417 | 0.08 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chr6_+_41021027 | 0.07 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr1_-_21620877 | 0.07 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr12_+_18891045 | 0.07 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr19_-_41942344 | 0.07 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chr16_+_30007524 | 0.07 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr17_-_7232585 | 0.07 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr8_+_87111059 | 0.07 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr18_-_60985914 | 0.07 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr3_-_49726486 | 0.07 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr3_+_63805017 | 0.07 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr17_+_79031415 | 0.07 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr19_-_3500635 | 0.06 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr11_-_45939374 | 0.06 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chrX_+_56259316 | 0.06 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr11_+_66790816 | 0.06 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr14_-_24911868 | 0.06 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr4_+_57371509 | 0.06 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr16_-_11350036 | 0.06 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr17_+_38474489 | 0.06 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr7_+_17414360 | 0.06 |
ENST00000419463.1
|
AC019117.1
|
AC019117.1 |
| chr2_+_172864490 | 0.06 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr15_+_76030311 | 0.06 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr16_+_30675654 | 0.06 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr3_-_61237050 | 0.06 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr3_+_184080790 | 0.06 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr6_+_43445261 | 0.05 |
ENST00000372444.2
ENST00000372445.5 ENST00000436109.2 ENST00000372454.2 ENST00000442878.2 ENST00000259751.1 ENST00000372452.1 ENST00000372449.1 |
TJAP1
|
tight junction associated protein 1 (peripheral) |
| chr8_-_144655141 | 0.05 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr20_-_22565101 | 0.05 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr21_+_44866471 | 0.05 |
ENST00000448049.1
|
LINC00319
|
long intergenic non-protein coding RNA 319 |
| chr1_+_101702417 | 0.05 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr11_+_58390132 | 0.05 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chrX_-_148713365 | 0.05 |
ENST00000511776.1
ENST00000507237.1 |
TMEM185A
|
transmembrane protein 185A |
| chr14_-_37051798 | 0.05 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr19_+_10397648 | 0.05 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr16_-_50715239 | 0.05 |
ENST00000330943.4
ENST00000300590.3 |
SNX20
|
sorting nexin 20 |
| chr13_-_34250861 | 0.05 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr12_+_57853918 | 0.05 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr17_-_73937116 | 0.05 |
ENST00000586717.1
ENST00000389570.4 ENST00000319129.5 |
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chr12_-_62585203 | 0.05 |
ENST00000551449.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr21_+_30968360 | 0.05 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr15_-_43863695 | 0.05 |
ENST00000439195.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr7_-_130080681 | 0.05 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr13_-_114107839 | 0.05 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr19_+_41856816 | 0.05 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr17_-_74533963 | 0.05 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr21_+_30503282 | 0.05 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_16062820 | 0.05 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr1_-_151119087 | 0.05 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr12_-_62586543 | 0.05 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr7_+_100450328 | 0.05 |
ENST00000540482.1
ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9
|
solute carrier family 12, member 9 |
| chr12_+_7055631 | 0.04 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr16_-_31147020 | 0.04 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr11_-_72145669 | 0.04 |
ENST00000543042.1
ENST00000294053.3 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr3_+_42695176 | 0.04 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr22_-_46373004 | 0.04 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr11_-_119252425 | 0.04 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr19_+_38794797 | 0.04 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr11_+_46402297 | 0.04 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_49056635 | 0.04 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr8_-_21988558 | 0.04 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr2_+_183943464 | 0.04 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr19_+_10397621 | 0.04 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr12_+_19358192 | 0.04 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_-_153917700 | 0.04 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr12_+_54519842 | 0.04 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr1_-_153521714 | 0.04 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr17_-_61996160 | 0.04 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chr15_+_89182178 | 0.04 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_64068116 | 0.04 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr16_+_31120429 | 0.04 |
ENST00000484226.2
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr2_-_175870085 | 0.04 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr14_-_36988882 | 0.04 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr17_+_38119216 | 0.04 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chr20_+_48884002 | 0.04 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr11_+_1860200 | 0.04 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr19_-_42721819 | 0.04 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr11_+_117947724 | 0.04 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr15_+_40733387 | 0.04 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr19_+_55795493 | 0.04 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr11_+_117947782 | 0.04 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr16_+_30662360 | 0.04 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr1_-_145076068 | 0.04 |
ENST00000369345.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr4_+_166300084 | 0.04 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr1_+_154975110 | 0.04 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr12_+_52203789 | 0.04 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr17_+_27055798 | 0.04 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr16_+_2546033 | 0.04 |
ENST00000564543.1
ENST00000434757.2 |
RP11-20I23.1
TBC1D24
|
Uncharacterized protein TBC1 domain family, member 24 |
| chr3_-_182880541 | 0.04 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr4_-_175443484 | 0.04 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr4_-_168155169 | 0.04 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_-_40857179 | 0.04 |
ENST00000558750.1
ENST00000561011.1 ENST00000560305.1 ENST00000559911.1 ENST00000558113.1 ENST00000559103.1 ENST00000558918.1 ENST00000558871.1 ENST00000358005.3 ENST00000416810.2 |
C15orf57
|
chromosome 15 open reading frame 57 |
| chr14_+_85994943 | 0.04 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr6_+_292253 | 0.04 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr22_+_46481861 | 0.04 |
ENST00000360737.3
|
FLJ27365
|
hsa-mir-4763 |
| chr3_-_50360192 | 0.04 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr4_-_40632605 | 0.04 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_-_473135 | 0.03 |
ENST00000342584.3
|
CTD-2228K2.5
|
Uncharacterized protein |
| chr7_+_76139833 | 0.03 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr7_+_76139741 | 0.03 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr19_+_53761545 | 0.03 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr8_-_145060593 | 0.03 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr2_+_103089756 | 0.03 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr4_-_68620053 | 0.03 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr10_+_135340859 | 0.03 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr15_+_40532058 | 0.03 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_-_132589601 | 0.03 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr19_-_42931567 | 0.03 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr17_+_79859985 | 0.03 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr5_-_134914673 | 0.03 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr19_-_10213335 | 0.03 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr1_-_40782938 | 0.03 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr1_+_154975258 | 0.03 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr8_+_96037205 | 0.03 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr11_-_72145641 | 0.03 |
ENST00000538039.1
ENST00000445069.2 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr11_-_45939565 | 0.03 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr12_+_53645870 | 0.03 |
ENST00000329548.4
|
MFSD5
|
major facilitator superfamily domain containing 5 |
| chr1_+_97187318 | 0.03 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr6_-_26018007 | 0.03 |
ENST00000244573.3
|
HIST1H1A
|
histone cluster 1, H1a |
| chr11_-_3147835 | 0.03 |
ENST00000525498.1
|
OSBPL5
|
oxysterol binding protein-like 5 |
| chr2_-_218766698 | 0.03 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr16_+_3162557 | 0.03 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr14_-_91526462 | 0.03 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr6_+_34204642 | 0.03 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr10_-_31320860 | 0.03 |
ENST00000436087.2
ENST00000442986.1 ENST00000413025.1 ENST00000452305.1 |
ZNF438
|
zinc finger protein 438 |
| chr3_+_52279737 | 0.03 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr11_-_72070206 | 0.03 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_+_228395755 | 0.03 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr17_+_47572647 | 0.03 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr7_+_143318020 | 0.03 |
ENST00000444908.2
ENST00000518791.1 ENST00000411497.2 |
FAM115C
|
family with sequence similarity 115, member C |
| chr20_-_45530365 | 0.03 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr11_-_72145426 | 0.03 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr15_-_43559055 | 0.03 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr1_-_38412683 | 0.03 |
ENST00000373024.3
ENST00000373023.2 |
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr1_+_151483855 | 0.03 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr2_+_242127924 | 0.03 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr11_-_128737259 | 0.03 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chrX_-_106960285 | 0.03 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr6_+_30294612 | 0.03 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr17_-_7297833 | 0.03 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr16_+_20911174 | 0.03 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr9_-_98784042 | 0.03 |
ENST00000412122.2
|
LINC00092
|
long intergenic non-protein coding RNA 92 |
| chrX_+_69642881 | 0.03 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr12_+_57849048 | 0.03 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr1_+_155911480 | 0.03 |
ENST00000368318.3
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4 |
| chr8_+_99439214 | 0.03 |
ENST00000287042.4
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr5_-_127418755 | 0.03 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr5_-_34043310 | 0.03 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr8_+_32405785 | 0.03 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chrX_-_134049233 | 0.03 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr5_+_140810132 | 0.03 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr1_+_15736359 | 0.03 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr1_-_55341551 | 0.03 |
ENST00000537443.1
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr17_-_61996136 | 0.03 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr5_-_88120083 | 0.03 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr14_+_75745477 | 0.03 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_-_129076151 | 0.03 |
ENST00000259241.6
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr2_+_183989157 | 0.03 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr15_+_89182156 | 0.03 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_-_82338460 | 0.03 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr16_+_640201 | 0.03 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr15_+_41099254 | 0.03 |
ENST00000570108.1
ENST00000564258.1 ENST00000355341.4 ENST00000336455.5 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr9_+_4662282 | 0.03 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr7_+_2687173 | 0.03 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr7_+_30791743 | 0.03 |
ENST00000013222.5
ENST00000409539.1 |
INMT
|
indolethylamine N-methyltransferase |
| chr19_-_11456872 | 0.03 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chrX_+_135618258 | 0.03 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr15_+_81489213 | 0.03 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr1_-_204329013 | 0.02 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0045013 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |