Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for TBX3
Z-value: 0.77
Transcription factors associated with TBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX3
|
ENSG00000135111.10 | T-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX3 | hg19_v2_chr12_-_115121962_115121987 | -0.54 | 2.7e-01 | Click! |
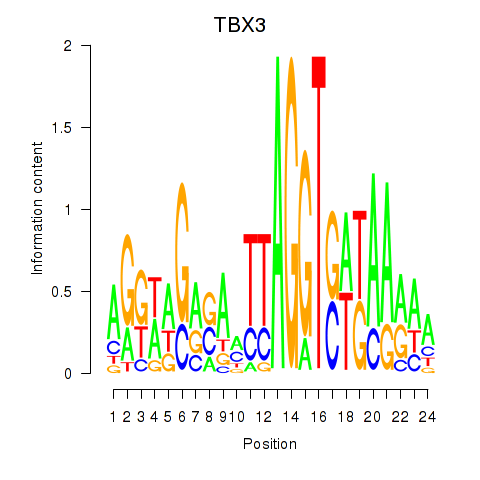
Activity profile of TBX3 motif
Sorted Z-values of TBX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 1.51 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_32821924 | 0.65 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_-_18258342 | 0.54 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr14_-_51863853 | 0.52 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr1_-_153085984 | 0.48 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr2_-_113594279 | 0.31 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_-_183560011 | 0.31 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr19_-_43835582 | 0.30 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr11_-_327537 | 0.27 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr20_+_34679725 | 0.27 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr11_+_327171 | 0.26 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr11_+_65082289 | 0.26 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr1_-_8075693 | 0.25 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr11_-_1619524 | 0.25 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr11_-_61849656 | 0.25 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr19_-_10679644 | 0.24 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr6_+_86195088 | 0.24 |
ENST00000437581.1
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr19_-_10679697 | 0.24 |
ENST00000335766.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr16_+_84801852 | 0.22 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr1_-_152552980 | 0.22 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr16_-_14726576 | 0.21 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr1_-_153283194 | 0.21 |
ENST00000290722.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr10_-_90611566 | 0.20 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr16_-_68057770 | 0.20 |
ENST00000332395.5
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 |
| chr1_-_119869846 | 0.20 |
ENST00000457719.1
|
RP11-418J17.3
|
RP11-418J17.3 |
| chr2_-_202298268 | 0.19 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr6_-_11382478 | 0.19 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_-_46068969 | 0.19 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr14_+_90422867 | 0.18 |
ENST00000553989.1
|
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr15_-_43212836 | 0.18 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr17_-_41132410 | 0.18 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr15_-_43212996 | 0.17 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr8_+_23386305 | 0.17 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr14_+_74416989 | 0.16 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr22_-_41258074 | 0.16 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr8_-_121825088 | 0.16 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr7_+_113091127 | 0.16 |
ENST00000433518.1
|
TSRM
|
Uncharacterized protein; Zinc finger domain-related protein TSRM |
| chr3_+_42897512 | 0.16 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr3_-_170626418 | 0.15 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr6_-_32821599 | 0.15 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr12_+_50478977 | 0.15 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_-_2883928 | 0.15 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr17_-_26220366 | 0.15 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr7_-_93519471 | 0.15 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr15_-_74501310 | 0.14 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr19_-_3478477 | 0.14 |
ENST00000591708.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr17_+_48351785 | 0.14 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr1_+_59522284 | 0.14 |
ENST00000449812.2
|
RP11-145M4.3
|
RP11-145M4.3 |
| chr8_-_81083341 | 0.14 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr11_+_34643600 | 0.14 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr19_+_55385731 | 0.13 |
ENST00000469767.1
ENST00000355524.3 ENST00000391725.3 ENST00000345937.4 ENST00000353758.4 ENST00000359272.4 ENST00000391723.3 ENST00000391724.3 |
FCAR
|
Fc fragment of IgA, receptor for |
| chr3_-_55539287 | 0.13 |
ENST00000472238.1
|
RP11-875H7.5
|
RP11-875H7.5 |
| chr11_+_71238313 | 0.13 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr4_+_71384300 | 0.13 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr6_-_55443975 | 0.12 |
ENST00000308161.4
ENST00000398661.2 ENST00000274901.4 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr7_+_102389434 | 0.12 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr7_+_95401877 | 0.12 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr7_-_76255444 | 0.12 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr12_+_48876275 | 0.11 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr3_+_63638372 | 0.11 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr5_+_40841410 | 0.11 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr3_+_52454971 | 0.11 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chrX_+_149887090 | 0.10 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr20_-_5931109 | 0.10 |
ENST00000203001.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr8_+_133931648 | 0.10 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr4_+_169575875 | 0.10 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_1800179 | 0.10 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr11_-_62607036 | 0.10 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr19_+_496454 | 0.10 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr1_+_159772121 | 0.10 |
ENST00000339348.5
ENST00000392235.3 ENST00000368106.3 |
FCRL6
|
Fc receptor-like 6 |
| chr6_+_32146268 | 0.10 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr18_-_5544241 | 0.10 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr8_+_86851932 | 0.10 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr15_-_59665062 | 0.10 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr6_+_31021225 | 0.09 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr1_+_171750776 | 0.09 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chr9_-_100954910 | 0.09 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr6_-_41254403 | 0.09 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr16_-_30381580 | 0.09 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr1_-_155959853 | 0.09 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr15_-_34635314 | 0.09 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr6_-_131277510 | 0.09 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr14_+_74111578 | 0.09 |
ENST00000554113.1
ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1
|
dynein, axonemal, light chain 1 |
| chrX_-_65259914 | 0.09 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr7_-_99381798 | 0.09 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr11_+_118889142 | 0.08 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr14_-_50087312 | 0.08 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like |
| chr5_+_148521046 | 0.08 |
ENST00000326685.7
ENST00000356541.3 ENST00000309868.7 |
ABLIM3
|
actin binding LIM protein family, member 3 |
| chrX_-_131623043 | 0.08 |
ENST00000421707.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr5_-_135290651 | 0.08 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr4_-_85654615 | 0.08 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr12_-_56121612 | 0.08 |
ENST00000546939.1
|
CD63
|
CD63 molecule |
| chr20_-_5931051 | 0.08 |
ENST00000453074.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr12_-_56120865 | 0.08 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr1_+_212208919 | 0.08 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr6_+_30852738 | 0.08 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_-_71532207 | 0.08 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_+_77924204 | 0.08 |
ENST00000555133.1
|
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr9_+_117904097 | 0.08 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr11_-_27722021 | 0.08 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr11_-_122932730 | 0.07 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr14_-_74416829 | 0.07 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr16_+_72088376 | 0.07 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr6_-_46703092 | 0.07 |
ENST00000541026.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_41132564 | 0.07 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr19_-_51531272 | 0.07 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr10_+_60028818 | 0.07 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr15_-_38852251 | 0.07 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr6_+_30853002 | 0.07 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_242612726 | 0.07 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr10_-_104262426 | 0.07 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr6_-_33385655 | 0.07 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_-_71293921 | 0.07 |
ENST00000398530.1
|
KRTAP5-11
|
keratin associated protein 5-11 |
| chr15_+_59665194 | 0.07 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr6_-_35888905 | 0.07 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_+_124125286 | 0.07 |
ENST00000368416.1
ENST00000368417.1 ENST00000546092.1 |
NKAIN2
|
Na+/K+ transporting ATPase interacting 2 |
| chr7_+_98476095 | 0.07 |
ENST00000359863.4
ENST00000355540.3 |
TRRAP
|
transformation/transcription domain-associated protein |
| chr11_+_71259466 | 0.07 |
ENST00000528743.2
|
KRTAP5-9
|
keratin associated protein 5-9 |
| chr10_+_51827648 | 0.07 |
ENST00000351071.6
ENST00000314664.7 ENST00000282633.5 |
FAM21A
|
family with sequence similarity 21, member A |
| chr5_+_139739772 | 0.07 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr18_+_5238055 | 0.07 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr22_+_24891210 | 0.07 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr6_-_55443831 | 0.07 |
ENST00000428842.1
ENST00000358072.5 ENST00000508459.1 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr19_+_39936207 | 0.06 |
ENST00000594729.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr4_+_40195262 | 0.06 |
ENST00000503941.1
|
RHOH
|
ras homolog family member H |
| chr19_-_42724261 | 0.06 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr10_+_94608218 | 0.06 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr20_-_1472053 | 0.06 |
ENST00000537284.1
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr15_+_77713299 | 0.06 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr19_-_51538118 | 0.06 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr5_-_39425222 | 0.06 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr9_+_104296243 | 0.06 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr17_+_45286387 | 0.06 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr12_+_7169887 | 0.06 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr2_-_24149918 | 0.06 |
ENST00000439915.1
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr11_-_122931881 | 0.06 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr19_-_51530916 | 0.06 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr14_+_21785693 | 0.06 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chrX_+_51942963 | 0.06 |
ENST00000375625.3
|
RP11-363G10.2
|
RP11-363G10.2 |
| chr3_-_128207349 | 0.06 |
ENST00000487848.1
|
GATA2
|
GATA binding protein 2 |
| chr4_-_183838747 | 0.06 |
ENST00000438320.2
|
DCTD
|
dCMP deaminase |
| chr14_+_70233810 | 0.06 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr22_-_38480100 | 0.06 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr19_-_42931567 | 0.06 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr15_+_59664884 | 0.06 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr17_-_28661065 | 0.06 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr10_-_71176623 | 0.06 |
ENST00000373306.4
|
TACR2
|
tachykinin receptor 2 |
| chr6_-_55443958 | 0.06 |
ENST00000370850.2
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr5_+_150040403 | 0.06 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr19_+_41257084 | 0.06 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr19_-_47735918 | 0.05 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr19_-_51531210 | 0.05 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr1_+_159770292 | 0.05 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr2_+_114647617 | 0.05 |
ENST00000536059.1
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr12_-_56120838 | 0.05 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr15_+_77712993 | 0.05 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr6_-_128222103 | 0.05 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr20_-_1472121 | 0.05 |
ENST00000444444.2
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr1_+_36396313 | 0.05 |
ENST00000324350.5
|
AGO3
|
argonaute RISC catalytic component 3 |
| chr4_-_108641608 | 0.05 |
ENST00000265174.4
|
PAPSS1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr1_+_32608566 | 0.05 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr3_+_133293278 | 0.05 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr15_+_74911430 | 0.05 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr17_+_43861680 | 0.05 |
ENST00000314537.5
|
CRHR1
|
corticotropin releasing hormone receptor 1 |
| chr15_-_74504597 | 0.05 |
ENST00000416286.3
|
STRA6
|
stimulated by retinoic acid 6 |
| chr12_-_8815404 | 0.05 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr15_+_40861487 | 0.05 |
ENST00000315616.7
ENST00000559271.1 |
RPUSD2
|
RNA pseudouridylate synthase domain containing 2 |
| chr20_-_44539538 | 0.05 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr4_-_69215699 | 0.05 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr19_-_16682987 | 0.05 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr2_+_242167319 | 0.05 |
ENST00000601871.1
|
AC104841.2
|
HCG1777198, isoform CRA_a; PRO2900; Uncharacterized protein |
| chr10_+_47894572 | 0.05 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr1_+_151020175 | 0.05 |
ENST00000368926.5
|
C1orf56
|
chromosome 1 open reading frame 56 |
| chr19_+_39936267 | 0.05 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr19_-_51538148 | 0.05 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr20_+_34359905 | 0.04 |
ENST00000374012.3
ENST00000439301.1 ENST00000339089.6 ENST00000374000.4 |
PHF20
|
PHD finger protein 20 |
| chr19_+_53517174 | 0.04 |
ENST00000602168.1
|
ERVV-1
|
endogenous retrovirus group V, member 1 |
| chr2_+_55459808 | 0.04 |
ENST00000404735.1
|
RPS27A
|
ribosomal protein S27a |
| chr1_+_172628154 | 0.04 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr8_-_20161466 | 0.04 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr19_+_39936186 | 0.04 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr2_+_216974020 | 0.04 |
ENST00000392132.2
ENST00000417391.1 |
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr7_-_86595190 | 0.04 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr11_-_11374904 | 0.04 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr1_+_36396791 | 0.04 |
ENST00000246314.6
|
AGO3
|
argonaute RISC catalytic component 3 |
| chr20_-_1472029 | 0.04 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr10_+_50507232 | 0.04 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr22_+_41487711 | 0.04 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr10_-_1071796 | 0.04 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr21_+_27011899 | 0.04 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr2_-_55459485 | 0.04 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr14_-_24911971 | 0.04 |
ENST00000555365.1
ENST00000399395.3 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr13_-_103719196 | 0.04 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr7_-_107204706 | 0.04 |
ENST00000393603.2
|
COG5
|
component of oligomeric golgi complex 5 |
| chr7_+_137761199 | 0.04 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr16_+_27078219 | 0.03 |
ENST00000418886.1
|
C16orf82
|
chromosome 16 open reading frame 82 |
| chr6_+_160542821 | 0.03 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr14_-_23446900 | 0.03 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr19_+_1077393 | 0.03 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr10_+_71561704 | 0.03 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.2 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.3 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 0.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.5 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.0 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |