Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
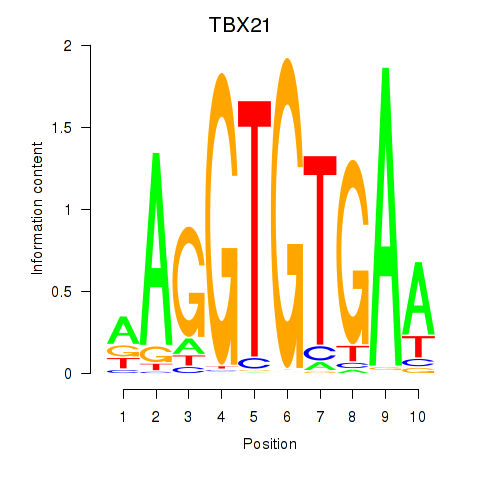
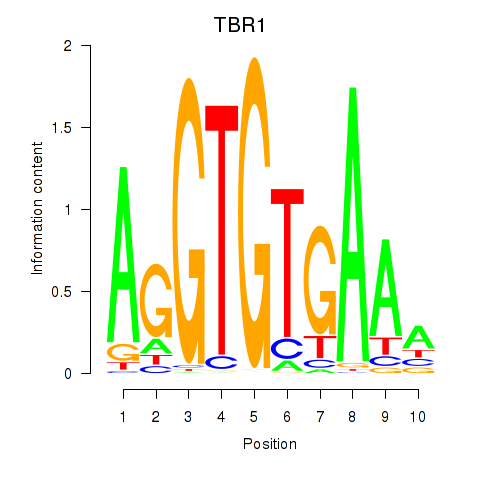
Results for TBX21_TBR1
Z-value: 0.70
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | T-box transcription factor 21 |
|
TBR1
|
ENSG00000136535.10 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | 0.01 | 9.8e-01 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_142315294 | 0.71 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr1_-_153029980 | 0.59 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr7_+_107220660 | 0.50 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_153066998 | 0.46 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr1_-_156786634 | 0.39 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr2_+_228678550 | 0.36 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr15_-_80263506 | 0.36 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr7_-_127032741 | 0.35 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr11_+_93754513 | 0.32 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr1_+_35734616 | 0.32 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr16_+_69139467 | 0.31 |
ENST00000569188.1
|
HAS3
|
hyaluronan synthase 3 |
| chr12_+_65672702 | 0.30 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr1_-_44482979 | 0.29 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr12_-_89746264 | 0.28 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr15_+_44119159 | 0.28 |
ENST00000263795.6
ENST00000381246.2 ENST00000452115.1 |
WDR76
|
WD repeat domain 76 |
| chr1_-_156786530 | 0.28 |
ENST00000368198.3
|
SH2D2A
|
SH2 domain containing 2A |
| chr1_+_10057274 | 0.27 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr9_+_26956371 | 0.27 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr12_-_52685312 | 0.26 |
ENST00000327741.5
|
KRT81
|
keratin 81 |
| chr11_+_64073699 | 0.24 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr17_-_76356148 | 0.24 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr19_-_7990991 | 0.23 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr19_+_10381769 | 0.23 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr18_-_59561417 | 0.23 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chr8_+_103876528 | 0.23 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr1_-_1009683 | 0.22 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr4_+_2626988 | 0.22 |
ENST00000509050.1
|
FAM193A
|
family with sequence similarity 193, member A |
| chrX_-_119693745 | 0.22 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr5_+_150040403 | 0.22 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr5_+_158654712 | 0.22 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr14_+_95078714 | 0.21 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr3_+_177534653 | 0.21 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr8_-_99955042 | 0.21 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr13_-_30683005 | 0.20 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr3_-_61237050 | 0.20 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr3_+_142315225 | 0.19 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr4_+_154622652 | 0.19 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr12_-_89746173 | 0.19 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr9_+_33750515 | 0.18 |
ENST00000361005.5
|
PRSS3
|
protease, serine, 3 |
| chr8_-_99954788 | 0.18 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr9_+_26956474 | 0.18 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr1_-_6260896 | 0.17 |
ENST00000497965.1
|
RPL22
|
ribosomal protein L22 |
| chr20_-_44516256 | 0.17 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr11_+_114271251 | 0.17 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr1_+_144989309 | 0.17 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr12_+_57854274 | 0.17 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr6_-_26108355 | 0.17 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr17_+_4046964 | 0.17 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr5_+_74807581 | 0.17 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr19_+_48949087 | 0.16 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr4_+_140222609 | 0.16 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chrX_+_105412290 | 0.16 |
ENST00000357175.2
ENST00000337685.2 |
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr18_+_55712915 | 0.16 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_+_32546145 | 0.16 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_+_74807886 | 0.16 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr14_+_21467414 | 0.16 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr2_-_157189180 | 0.15 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr14_+_77582905 | 0.15 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr16_+_53133070 | 0.15 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_155099927 | 0.15 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr4_+_74735102 | 0.15 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr19_+_56653064 | 0.15 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr1_-_205290865 | 0.15 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr14_-_62217779 | 0.15 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr1_-_108743471 | 0.15 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_+_76101379 | 0.14 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr16_+_50313426 | 0.14 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr6_-_13487825 | 0.14 |
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr13_-_30160925 | 0.14 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_+_144766617 | 0.14 |
ENST00000454097.1
ENST00000534303.1 ENST00000529833.1 ENST00000530574.1 ENST00000442058.2 ENST00000358656.4 ENST00000526970.1 ENST00000532158.1 |
ZNF707
|
zinc finger protein 707 |
| chr16_+_30386098 | 0.14 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr7_-_120497178 | 0.13 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr10_-_75226166 | 0.13 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr11_-_76155700 | 0.13 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr9_+_33750667 | 0.13 |
ENST00000457896.1
ENST00000342836.4 ENST00000429677.3 |
PRSS3
|
protease, serine, 3 |
| chr5_+_137514403 | 0.13 |
ENST00000513276.1
|
KIF20A
|
kinesin family member 20A |
| chr12_+_66582919 | 0.13 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr22_-_38699003 | 0.13 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr19_+_1000418 | 0.13 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr3_-_197024394 | 0.13 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_102758210 | 0.12 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr11_-_615570 | 0.12 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr2_+_102758751 | 0.12 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr6_-_160166218 | 0.12 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr1_+_156105878 | 0.12 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr16_-_88717482 | 0.12 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr5_+_132009675 | 0.12 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr12_+_32112340 | 0.12 |
ENST00000540924.1
ENST00000312561.4 |
KIAA1551
|
KIAA1551 |
| chr16_+_69140122 | 0.12 |
ENST00000219322.3
|
HAS3
|
hyaluronan synthase 3 |
| chr11_-_18956556 | 0.11 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr1_-_44818599 | 0.11 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr11_+_114270752 | 0.11 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr19_-_35068562 | 0.11 |
ENST00000595013.1
|
SCGB1B2P
|
secretoglobin, family 1B, member 2, pseudogene |
| chr1_+_28586006 | 0.11 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr14_+_32546274 | 0.11 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_115124257 | 0.11 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr2_-_61697862 | 0.11 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr9_-_96717654 | 0.10 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr16_-_88717423 | 0.10 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr10_+_4868460 | 0.10 |
ENST00000532248.1
ENST00000345253.5 ENST00000334019.4 |
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr17_-_38721711 | 0.10 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr19_+_38779778 | 0.10 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chrX_-_15872914 | 0.10 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_207630081 | 0.10 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr2_-_230933709 | 0.10 |
ENST00000436869.1
ENST00000295190.4 |
SLC16A14
|
solute carrier family 16, member 14 |
| chrX_+_16804544 | 0.09 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr11_-_60010556 | 0.09 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr6_-_130536774 | 0.09 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr3_-_57678772 | 0.09 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr8_+_120220561 | 0.09 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr19_+_50529329 | 0.09 |
ENST00000599155.1
|
ZNF473
|
zinc finger protein 473 |
| chr9_+_17135016 | 0.09 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr1_+_202172848 | 0.09 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr16_-_4395905 | 0.09 |
ENST00000571941.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr12_+_65672423 | 0.09 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr9_+_34329493 | 0.09 |
ENST00000379158.2
ENST00000346365.4 ENST00000379155.5 |
NUDT2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr19_+_3762703 | 0.09 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr4_+_48833070 | 0.09 |
ENST00000511662.1
ENST00000508996.1 ENST00000507210.1 ENST00000264312.7 ENST00000396448.2 ENST00000512236.1 ENST00000509164.1 ENST00000511102.1 ENST00000381473.3 |
OCIAD1
|
OCIA domain containing 1 |
| chr17_+_26662679 | 0.09 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chrX_-_132549506 | 0.09 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr20_+_58630972 | 0.09 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr19_-_50529193 | 0.09 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr19_-_36236292 | 0.09 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr13_+_76378305 | 0.08 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr3_+_57741957 | 0.08 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr6_+_131894284 | 0.08 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr2_+_201994042 | 0.08 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr17_-_72619869 | 0.08 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr4_+_48833312 | 0.08 |
ENST00000508293.1
ENST00000513391.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr16_+_8715536 | 0.08 |
ENST00000563958.1
ENST00000381920.3 ENST00000564554.1 |
METTL22
|
methyltransferase like 22 |
| chr3_-_107777208 | 0.08 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr20_-_1165319 | 0.08 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr19_-_47226468 | 0.08 |
ENST00000600615.1
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr2_-_113594279 | 0.08 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_+_19967014 | 0.08 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr20_+_2517253 | 0.08 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr5_+_133861339 | 0.08 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr11_-_95657231 | 0.08 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr7_-_87849340 | 0.08 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr8_+_22428457 | 0.08 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr17_-_43502987 | 0.08 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr11_-_64052111 | 0.08 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr6_-_110500826 | 0.08 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr4_+_108746282 | 0.08 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_-_113247543 | 0.08 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr3_+_185000729 | 0.08 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr4_+_48833015 | 0.07 |
ENST00000509122.1
ENST00000509664.1 ENST00000505922.2 ENST00000514981.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr22_+_42196666 | 0.07 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr6_-_97731019 | 0.07 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr19_-_43702231 | 0.07 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr17_+_8191815 | 0.07 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr2_+_29033682 | 0.07 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr19_-_3500635 | 0.07 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr1_+_202995611 | 0.07 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr14_-_98444386 | 0.07 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr5_-_49737184 | 0.07 |
ENST00000508934.1
ENST00000303221.5 |
EMB
|
embigin |
| chr1_+_38022572 | 0.07 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chrX_-_19689106 | 0.07 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_152146385 | 0.07 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chrX_+_47077680 | 0.07 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr8_+_145218673 | 0.07 |
ENST00000326134.5
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr9_+_35829208 | 0.07 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr12_-_71148413 | 0.07 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr9_-_117853297 | 0.07 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr12_+_21525818 | 0.07 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr3_+_167453493 | 0.07 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr7_+_133261209 | 0.07 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr1_+_93544791 | 0.07 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr11_-_28129656 | 0.07 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr9_+_78505581 | 0.07 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr11_+_65687158 | 0.07 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr4_+_718896 | 0.07 |
ENST00000433814.1
|
PCGF3
|
polycomb group ring finger 3 |
| chr19_-_39390212 | 0.07 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr2_+_162087577 | 0.07 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr15_-_43559055 | 0.07 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr11_-_82997013 | 0.06 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr16_+_30212378 | 0.06 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_-_60005365 | 0.06 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr3_+_112709804 | 0.06 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr1_+_154975110 | 0.06 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr3_+_187896331 | 0.06 |
ENST00000392468.2
|
AC022498.1
|
Uncharacterized protein |
| chr9_+_134378289 | 0.06 |
ENST00000423007.1
ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1
|
protein-O-mannosyltransferase 1 |
| chr4_+_87857538 | 0.06 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr16_-_18911366 | 0.06 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_+_73106926 | 0.06 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr6_-_24646249 | 0.06 |
ENST00000430948.2
ENST00000537886.1 ENST00000535378.1 ENST00000378214.3 |
KIAA0319
|
KIAA0319 |
| chr15_-_65903512 | 0.06 |
ENST00000567923.1
|
VWA9
|
von Willebrand factor A domain containing 9 |
| chr10_+_135050908 | 0.06 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chrX_-_128788914 | 0.06 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr2_+_219246746 | 0.06 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr19_-_55580838 | 0.06 |
ENST00000396247.3
ENST00000586945.1 ENST00000587026.1 |
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr12_+_109490370 | 0.06 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr19_-_44174330 | 0.06 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_+_46185111 | 0.06 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr19_-_55549624 | 0.06 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr3_+_124355944 | 0.06 |
ENST00000459915.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr20_+_30193083 | 0.06 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr1_-_114414316 | 0.06 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr15_+_89922345 | 0.06 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr2_+_58134756 | 0.06 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.4 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1903660 | cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.4 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.0 | GO:0051714 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) natural killer cell tolerance induction(GO:0002519) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0002001 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0002645 | positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 1.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |