Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
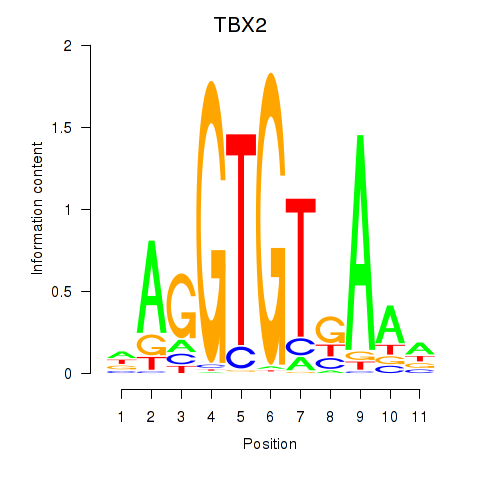
Results for TBX2
Z-value: 0.62
Transcription factors associated with TBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX2
|
ENSG00000121068.9 | T-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX2 | hg19_v2_chr17_+_59477233_59477263 | -0.51 | 3.0e-01 | Click! |
Activity profile of TBX2 motif
Sorted Z-values of TBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_103876528 | 0.43 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr6_-_110500826 | 0.42 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr1_+_35734616 | 0.40 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr14_+_21467414 | 0.40 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr7_+_107220899 | 0.40 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_+_142315294 | 0.36 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr7_+_107220660 | 0.34 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr11_-_18956556 | 0.32 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr2_+_29033682 | 0.32 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr2_+_102758271 | 0.31 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr5_-_138780159 | 0.27 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr19_-_43702231 | 0.24 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr17_-_60005365 | 0.24 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr1_-_183387723 | 0.23 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr7_-_108209897 | 0.23 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr8_-_99954788 | 0.22 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr3_+_121554046 | 0.22 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr3_+_142315225 | 0.21 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr3_-_194207388 | 0.21 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr11_-_112034831 | 0.21 |
ENST00000280357.7
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr12_+_65672702 | 0.20 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr20_-_45530365 | 0.20 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr11_+_66790816 | 0.19 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr1_+_144989309 | 0.19 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr17_+_46185111 | 0.19 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr8_-_27457494 | 0.17 |
ENST00000521770.1
|
CLU
|
clusterin |
| chr4_-_89619386 | 0.17 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr7_-_127032741 | 0.17 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr16_-_4303767 | 0.17 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr2_+_189156586 | 0.16 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_-_169769787 | 0.16 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr11_-_111649074 | 0.16 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr10_+_135050908 | 0.16 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr19_+_50832943 | 0.16 |
ENST00000542413.1
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr9_+_114287433 | 0.16 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr9_+_26956474 | 0.16 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr20_+_34020827 | 0.15 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr2_+_102758210 | 0.15 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr3_+_129207033 | 0.15 |
ENST00000507221.1
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr22_-_29138386 | 0.15 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr5_-_64858944 | 0.14 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chrX_-_119693745 | 0.14 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr2_+_102758751 | 0.14 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr2_-_105953912 | 0.14 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr15_-_55581954 | 0.14 |
ENST00000336787.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_12849073 | 0.14 |
ENST00000332427.2
ENST00000540796.1 |
GPR19
|
G protein-coupled receptor 19 |
| chr17_-_36884451 | 0.14 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr12_+_32112340 | 0.14 |
ENST00000540924.1
ENST00000312561.4 |
KIAA1551
|
KIAA1551 |
| chr1_-_205391178 | 0.14 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr19_+_50832895 | 0.13 |
ENST00000600355.1
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr8_+_145490549 | 0.13 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr4_-_186732048 | 0.13 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_69373210 | 0.13 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr14_+_21525981 | 0.13 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr14_-_45603657 | 0.13 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr6_-_105584560 | 0.13 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr15_-_52030293 | 0.13 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr11_+_20409070 | 0.13 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr11_+_122709200 | 0.12 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_-_71751715 | 0.12 |
ENST00000535947.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr9_-_35042824 | 0.12 |
ENST00000595331.1
|
FLJ00273
|
FLJ00273 |
| chr1_+_38022572 | 0.12 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr6_-_97731019 | 0.12 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr11_+_114271251 | 0.12 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr14_+_32546274 | 0.12 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_-_15165854 | 0.12 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr12_+_11905413 | 0.11 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr14_-_55658323 | 0.11 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr22_-_51001332 | 0.11 |
ENST00000406915.3
|
SYCE3
|
synaptonemal complex central element protein 3 |
| chr4_-_4249924 | 0.11 |
ENST00000254742.2
ENST00000382753.4 ENST00000540397.1 ENST00000538516.1 |
TMEM128
|
transmembrane protein 128 |
| chr3_-_197024394 | 0.11 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_30212378 | 0.11 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chrX_-_119709637 | 0.11 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr14_+_32546145 | 0.11 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr13_-_48669232 | 0.11 |
ENST00000258648.2
ENST00000378586.1 |
MED4
|
mediator complex subunit 4 |
| chr16_+_16481306 | 0.11 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr22_+_40441456 | 0.11 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chr3_-_119379719 | 0.11 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr1_-_52456352 | 0.10 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr12_+_65672423 | 0.10 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr11_-_114271139 | 0.10 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr8_-_124428569 | 0.10 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr9_+_40028620 | 0.10 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr11_-_16430399 | 0.10 |
ENST00000528252.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_205744205 | 0.10 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr7_-_48019101 | 0.10 |
ENST00000432627.1
ENST00000258774.5 ENST00000432325.1 ENST00000446009.1 |
HUS1
|
HUS1 checkpoint homolog (S. pombe) |
| chr2_+_53994927 | 0.10 |
ENST00000295304.4
|
CHAC2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chr9_+_26956371 | 0.10 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr2_-_75796837 | 0.10 |
ENST00000233712.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr7_+_108210012 | 0.09 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr17_-_60005329 | 0.09 |
ENST00000251334.6
|
INTS2
|
integrator complex subunit 2 |
| chr7_+_114562172 | 0.09 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr5_+_137514403 | 0.09 |
ENST00000513276.1
|
KIF20A
|
kinesin family member 20A |
| chr19_+_46000506 | 0.09 |
ENST00000396737.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr1_+_172628154 | 0.09 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr4_-_153700864 | 0.09 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr11_+_114270752 | 0.09 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr9_+_127023704 | 0.09 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr19_-_41942344 | 0.09 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chr3_+_32726774 | 0.09 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr2_+_201994042 | 0.09 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr14_-_55658252 | 0.09 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr3_+_112930387 | 0.09 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr8_-_90769422 | 0.09 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr20_-_1165117 | 0.09 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr7_-_7782204 | 0.09 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr14_+_77582905 | 0.09 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr9_-_134585221 | 0.08 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr14_-_35183755 | 0.08 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr11_-_28129656 | 0.08 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chrX_-_53461288 | 0.08 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_-_68628543 | 0.08 |
ENST00000396496.2
ENST00000511257.1 ENST00000383374.2 |
CCDC125
|
coiled-coil domain containing 125 |
| chr17_+_30771279 | 0.08 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr19_-_44405941 | 0.08 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr2_+_120770581 | 0.08 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr4_+_140222609 | 0.08 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr4_+_141445333 | 0.08 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr19_+_52264449 | 0.08 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr2_-_148778323 | 0.08 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr3_+_183967409 | 0.08 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chrX_-_53461305 | 0.08 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chrX_+_105412290 | 0.08 |
ENST00000357175.2
ENST00000337685.2 |
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr6_-_33679452 | 0.08 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr9_+_33750515 | 0.08 |
ENST00000361005.5
|
PRSS3
|
protease, serine, 3 |
| chr3_+_4535155 | 0.08 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_-_37407172 | 0.08 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr12_-_57443886 | 0.08 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr3_+_185431080 | 0.08 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr5_+_110559312 | 0.08 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr1_+_100111479 | 0.08 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr1_-_245026388 | 0.08 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr3_-_61237050 | 0.08 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr1_-_150780757 | 0.08 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr12_-_8815404 | 0.07 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr6_+_83777374 | 0.07 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr6_-_86099898 | 0.07 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr11_+_111412271 | 0.07 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr22_+_18593097 | 0.07 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr3_+_184016986 | 0.07 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr1_+_178482262 | 0.07 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr19_+_19779589 | 0.07 |
ENST00000541458.1
|
ZNF101
|
zinc finger protein 101 |
| chr1_-_6260896 | 0.07 |
ENST00000497965.1
|
RPL22
|
ribosomal protein L22 |
| chr1_+_89829610 | 0.07 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr12_-_111180644 | 0.07 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr19_-_44952635 | 0.07 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr9_+_17135016 | 0.07 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr11_+_19138670 | 0.07 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr22_-_42342733 | 0.07 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr14_-_74025625 | 0.07 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr2_+_179184955 | 0.07 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr11_+_107799118 | 0.07 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr2_-_136743169 | 0.07 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr19_-_39523165 | 0.07 |
ENST00000509137.2
ENST00000292853.4 |
FBXO27
|
F-box protein 27 |
| chr18_-_48723129 | 0.07 |
ENST00000592416.1
|
MEX3C
|
mex-3 RNA binding family member C |
| chr7_+_16700806 | 0.07 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_15104040 | 0.07 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr7_+_99102267 | 0.07 |
ENST00000326775.5
ENST00000451158.1 |
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr12_+_124155652 | 0.07 |
ENST00000426174.2
ENST00000303372.5 |
TCTN2
|
tectonic family member 2 |
| chr11_-_92931098 | 0.07 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr1_-_46598371 | 0.07 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr5_-_102898465 | 0.07 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr15_+_89922345 | 0.07 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr6_-_84937314 | 0.07 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr13_+_98628886 | 0.07 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr1_+_19967014 | 0.07 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr1_+_201708992 | 0.07 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr11_-_85779971 | 0.06 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_-_23446900 | 0.06 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr12_+_19358228 | 0.06 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr5_-_131132614 | 0.06 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr11_+_28129795 | 0.06 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr17_+_35849937 | 0.06 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr1_+_100111580 | 0.06 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr4_-_2243839 | 0.06 |
ENST00000511885.2
ENST00000506763.1 ENST00000514395.1 ENST00000502440.1 ENST00000243706.4 ENST00000443786.2 |
POLN
HAUS3
|
polymerase (DNA directed) nu HAUS augmin-like complex, subunit 3 |
| chr6_-_34639733 | 0.06 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr4_-_185303418 | 0.06 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr20_+_816695 | 0.06 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr17_+_19314432 | 0.06 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr11_+_129685707 | 0.06 |
ENST00000281441.3
|
TMEM45B
|
transmembrane protein 45B |
| chr9_-_103115135 | 0.06 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr12_+_56660633 | 0.06 |
ENST00000308197.5
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr2_+_219247021 | 0.06 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr15_+_67547113 | 0.06 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr3_+_112709804 | 0.06 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr2_-_103353277 | 0.06 |
ENST00000258436.5
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr4_+_26322185 | 0.06 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_-_55290773 | 0.06 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr2_-_39664405 | 0.06 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr6_-_26124138 | 0.06 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr20_-_271304 | 0.06 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr20_+_25388293 | 0.06 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr17_+_28804380 | 0.05 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr15_-_90294523 | 0.05 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chr9_+_123837141 | 0.05 |
ENST00000373865.2
|
CNTRL
|
centriolin |
| chr3_+_137728842 | 0.05 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr19_-_40772221 | 0.05 |
ENST00000441941.2
ENST00000580747.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chrX_+_16804544 | 0.05 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr7_+_17414360 | 0.05 |
ENST00000419463.1
|
AC019117.1
|
AC019117.1 |
| chr9_+_131452239 | 0.05 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.3 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0003163 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) |
| 0.0 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0060971 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.0 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 0.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |