Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
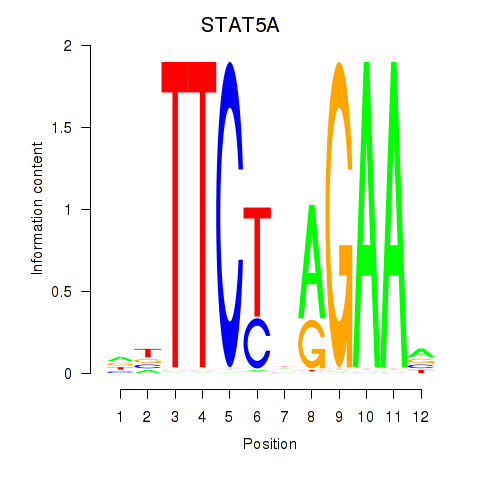
Results for STAT5A
Z-value: 0.86
Transcription factors associated with STAT5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5A
|
ENSG00000126561.12 | signal transducer and activator of transcription 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5A | hg19_v2_chr17_+_40440481_40440561 | -0.15 | 7.7e-01 | Click! |
Activity profile of STAT5A motif
Sorted Z-values of STAT5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_133656995 | 0.64 |
ENST00000356456.5
|
ZNF140
|
zinc finger protein 140 |
| chr20_+_58571419 | 0.59 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr13_+_20268547 | 0.51 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr8_-_123139423 | 0.43 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr3_-_165555200 | 0.41 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr3_-_4927447 | 0.38 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr16_+_70207686 | 0.38 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr10_+_6625605 | 0.38 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr2_+_181988620 | 0.38 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr11_-_111320706 | 0.38 |
ENST00000531398.1
|
POU2AF1
|
POU class 2 associating factor 1 |
| chr7_+_30589829 | 0.37 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr7_-_139727118 | 0.36 |
ENST00000484111.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr15_+_63414760 | 0.35 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chrX_+_106045891 | 0.35 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr12_+_66696322 | 0.35 |
ENST00000247815.4
|
HELB
|
helicase (DNA) B |
| chr4_+_130017268 | 0.34 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr12_+_21207503 | 0.31 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_75796837 | 0.31 |
ENST00000233712.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr3_-_160116995 | 0.30 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_-_71750865 | 0.30 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr10_-_33625154 | 0.30 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr12_-_15082050 | 0.29 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr1_-_115632035 | 0.29 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr11_-_114271139 | 0.27 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr6_+_32605134 | 0.27 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr3_-_160117301 | 0.27 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr1_-_226065330 | 0.27 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr7_+_142829162 | 0.27 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr2_-_98972468 | 0.26 |
ENST00000454230.1
|
AC092675.3
|
Uncharacterized protein |
| chr1_-_55230165 | 0.25 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr14_-_51135005 | 0.25 |
ENST00000556735.1
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr6_+_32605195 | 0.24 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr8_+_101349823 | 0.24 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr6_-_56716686 | 0.24 |
ENST00000520645.1
|
DST
|
dystonin |
| chr2_+_149894968 | 0.24 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr10_+_90484301 | 0.24 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr3_-_139108463 | 0.24 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr17_+_61151306 | 0.23 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_-_160117035 | 0.23 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr2_-_55237484 | 0.23 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr11_-_111649074 | 0.23 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_-_8815299 | 0.22 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr2_+_149895207 | 0.22 |
ENST00000409876.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr9_-_89562104 | 0.21 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr18_-_64271316 | 0.21 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr17_-_72968837 | 0.21 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr2_-_165630264 | 0.21 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr4_+_38511367 | 0.21 |
ENST00000507056.1
|
RP11-213G21.1
|
RP11-213G21.1 |
| chr4_+_108815402 | 0.21 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_+_158975744 | 0.21 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr8_+_109455830 | 0.20 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr4_+_25378912 | 0.20 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr20_+_33464368 | 0.20 |
ENST00000484354.1
ENST00000493805.2 ENST00000473172.1 |
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr3_-_78719376 | 0.20 |
ENST00000495961.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr13_-_51101468 | 0.19 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr5_+_127039075 | 0.19 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr2_+_37423618 | 0.19 |
ENST00000402297.1
ENST00000397064.2 ENST00000406711.1 ENST00000392061.2 ENST00000397226.2 |
AC007390.5
|
CEBPZ antisense RNA 1 |
| chr17_-_34207295 | 0.19 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr19_-_44809121 | 0.19 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr19_-_19729725 | 0.19 |
ENST00000251203.9
|
PBX4
|
pre-B-cell leukemia homeobox 4 |
| chr1_+_59775752 | 0.19 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr14_-_24977457 | 0.18 |
ENST00000250378.3
ENST00000206446.4 |
CMA1
|
chymase 1, mast cell |
| chr19_-_37407172 | 0.18 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr20_+_58251716 | 0.18 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr12_+_79357815 | 0.18 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr17_+_67498396 | 0.18 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_+_133657461 | 0.18 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr19_-_44405941 | 0.17 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr12_+_117013656 | 0.17 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr17_-_60005365 | 0.17 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr2_-_203735586 | 0.17 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr11_-_82997371 | 0.17 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr12_+_44152740 | 0.17 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr13_-_99404875 | 0.17 |
ENST00000376503.5
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr1_-_245026388 | 0.17 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr1_-_197115818 | 0.17 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr3_+_142342228 | 0.17 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr8_-_33330595 | 0.17 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr7_-_48019101 | 0.16 |
ENST00000432627.1
ENST00000258774.5 ENST00000432325.1 ENST00000446009.1 |
HUS1
|
HUS1 checkpoint homolog (S. pombe) |
| chr2_+_163200848 | 0.16 |
ENST00000233612.4
|
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr18_+_61575200 | 0.16 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr1_+_38478432 | 0.16 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr10_-_5652705 | 0.16 |
ENST00000425246.1
|
RP11-336A10.5
|
RP11-336A10.5 |
| chr20_+_5987890 | 0.16 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr6_+_116892641 | 0.16 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chr18_-_66382289 | 0.16 |
ENST00000443099.2
ENST00000562706.1 ENST00000544714.2 |
TMX3
|
thioredoxin-related transmembrane protein 3 |
| chr18_-_68004529 | 0.16 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr14_-_74025625 | 0.16 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr19_+_37960240 | 0.16 |
ENST00000388801.3
|
ZNF570
|
zinc finger protein 570 |
| chr4_-_146019693 | 0.16 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr10_-_6622201 | 0.16 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr19_+_57106624 | 0.16 |
ENST00000599599.1
|
ZNF71
|
zinc finger protein 71 |
| chr9_+_86237963 | 0.15 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr12_+_69080734 | 0.15 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr9_+_114287433 | 0.15 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr7_+_99425633 | 0.15 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr2_+_112813134 | 0.15 |
ENST00000452614.1
|
TMEM87B
|
transmembrane protein 87B |
| chr14_+_39583427 | 0.15 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr19_-_44952635 | 0.15 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr1_-_48937821 | 0.15 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr6_+_122793058 | 0.15 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr14_-_36983034 | 0.15 |
ENST00000518529.2
|
SFTA3
|
surfactant associated 3 |
| chr14_+_90422239 | 0.15 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr20_-_52612705 | 0.14 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_-_48937682 | 0.14 |
ENST00000371843.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr17_+_76356516 | 0.14 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr7_-_42971759 | 0.14 |
ENST00000538645.1
ENST00000445517.1 ENST00000223321.4 |
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr18_+_61144160 | 0.14 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr11_+_114271367 | 0.14 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr3_-_171489085 | 0.14 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr19_+_52430688 | 0.14 |
ENST00000391794.4
ENST00000599683.1 ENST00000600853.1 |
ZNF613
|
zinc finger protein 613 |
| chr2_-_68290106 | 0.14 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr11_+_114271251 | 0.14 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr17_+_32582293 | 0.14 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr2_+_238969728 | 0.14 |
ENST00000416757.1
|
SCLY
|
selenocysteine lyase |
| chr14_+_45605127 | 0.14 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr3_+_160117418 | 0.14 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr14_-_21270561 | 0.14 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr6_+_76330355 | 0.14 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr19_+_44669221 | 0.14 |
ENST00000590578.1
ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226
|
zinc finger protein 226 |
| chr17_-_60005329 | 0.14 |
ENST00000251334.6
|
INTS2
|
integrator complex subunit 2 |
| chr1_-_115301235 | 0.14 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_+_22591276 | 0.14 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr1_-_182558374 | 0.14 |
ENST00000367559.3
ENST00000539397.1 |
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
| chr14_-_94759595 | 0.14 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr7_-_130597935 | 0.13 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr19_+_12203069 | 0.13 |
ENST00000430298.2
ENST00000339302.4 |
ZNF788
ZNF788
|
zinc finger family member 788 Zinc finger protein 788 |
| chr5_-_42812143 | 0.13 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_-_45004556 | 0.13 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chrX_-_3264682 | 0.13 |
ENST00000217939.6
|
MXRA5
|
matrix-remodelling associated 5 |
| chr16_+_16481306 | 0.13 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr22_+_21128167 | 0.13 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr17_+_6347761 | 0.13 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr21_+_34602377 | 0.13 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr19_+_58193337 | 0.13 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr17_+_48823975 | 0.13 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr8_-_27469383 | 0.13 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr8_+_67104940 | 0.13 |
ENST00000517689.1
|
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr4_+_86396321 | 0.13 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_+_6544328 | 0.13 |
ENST00000570330.1
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr12_-_21910775 | 0.13 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr1_-_94344686 | 0.13 |
ENST00000528680.1
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr14_-_100841930 | 0.13 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr5_+_112312454 | 0.13 |
ENST00000543319.1
|
DCP2
|
decapping mRNA 2 |
| chr2_+_196522032 | 0.13 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr20_+_19738792 | 0.13 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr12_+_28605426 | 0.13 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_20134713 | 0.13 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2 |
| chr11_+_73358594 | 0.13 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr11_+_36589547 | 0.13 |
ENST00000299440.5
|
RAG1
|
recombination activating gene 1 |
| chr8_-_102803163 | 0.13 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr3_-_93692681 | 0.13 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr19_+_44617511 | 0.13 |
ENST00000262894.6
ENST00000588926.1 ENST00000592780.1 |
ZNF225
|
zinc finger protein 225 |
| chr19_+_8274512 | 0.13 |
ENST00000560412.1
|
CERS4
|
ceramide synthase 4 |
| chr9_-_94877658 | 0.13 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr14_+_63671105 | 0.13 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr1_-_144994840 | 0.12 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_158969752 | 0.12 |
ENST00000566111.1
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_+_173763250 | 0.12 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr3_+_141144963 | 0.12 |
ENST00000510726.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_+_114271314 | 0.12 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr1_-_144994909 | 0.12 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_+_37407351 | 0.12 |
ENST00000455427.2
ENST00000587857.1 |
ZNF568
|
zinc finger protein 568 |
| chr2_+_112812778 | 0.12 |
ENST00000283206.4
|
TMEM87B
|
transmembrane protein 87B |
| chr4_-_38807199 | 0.12 |
ENST00000508364.1
|
TLR1
|
toll-like receptor 1 |
| chr8_-_27468717 | 0.12 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr8_-_103884640 | 0.12 |
ENST00000520402.1
|
AZIN1
|
antizyme inhibitor 1 |
| chr3_+_122296443 | 0.12 |
ENST00000464300.2
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr16_+_30996502 | 0.12 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr9_+_86238016 | 0.12 |
ENST00000530832.1
ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr11_-_104893863 | 0.12 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr4_-_74486109 | 0.12 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_+_121163466 | 0.12 |
ENST00000527762.1
ENST00000534230.1 ENST00000392789.2 |
SC5D
|
sterol-C5-desaturase |
| chr6_+_123038689 | 0.12 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_-_5004241 | 0.12 |
ENST00000319533.5
ENST00000380051.2 |
RPP40
|
ribonuclease P/MRP 40kDa subunit |
| chr12_+_10658489 | 0.12 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr15_-_65282274 | 0.12 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr12_-_62997214 | 0.12 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr11_+_114270752 | 0.12 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr14_-_92333873 | 0.12 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chr7_+_5938351 | 0.12 |
ENST00000325974.6
|
CCZ1
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr15_-_65282232 | 0.12 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr19_+_7953417 | 0.12 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr9_-_25678856 | 0.11 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr10_-_116444371 | 0.11 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr9_-_75653627 | 0.11 |
ENST00000446946.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chrX_-_41449204 | 0.11 |
ENST00000378179.3
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr15_+_50474385 | 0.11 |
ENST00000267842.5
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr11_-_85430356 | 0.11 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_142558078 | 0.11 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr19_+_44507091 | 0.11 |
ENST00000429154.2
ENST00000585632.1 |
ZNF230
|
zinc finger protein 230 |
| chr14_-_58893832 | 0.11 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr6_-_86303523 | 0.11 |
ENST00000513865.1
ENST00000369627.2 ENST00000514419.1 ENST00000509338.1 ENST00000314673.3 ENST00000346348.3 |
SNX14
|
sorting nexin 14 |
| chr4_-_156298087 | 0.11 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr2_-_10830093 | 0.11 |
ENST00000381685.5
ENST00000345985.3 ENST00000542668.1 ENST00000538384.1 |
NOL10
|
nucleolar protein 10 |
| chr11_+_58912240 | 0.11 |
ENST00000527629.1
ENST00000361723.3 ENST00000531408.1 |
FAM111A
|
family with sequence similarity 111, member A |
| chr3_-_160823158 | 0.11 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr11_+_19138670 | 0.11 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr17_-_39459103 | 0.11 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr19_-_41220540 | 0.11 |
ENST00000594490.1
|
ADCK4
|
aarF domain containing kinase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.2 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.3 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.2 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) sensory neuron axon guidance(GO:0097374) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.4 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.3 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.2 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.3 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.3 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:1903060 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0070943 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.0 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.6 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.4 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.2 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.5 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |