Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
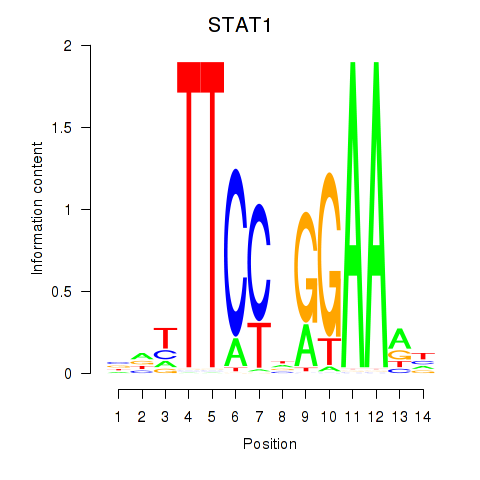
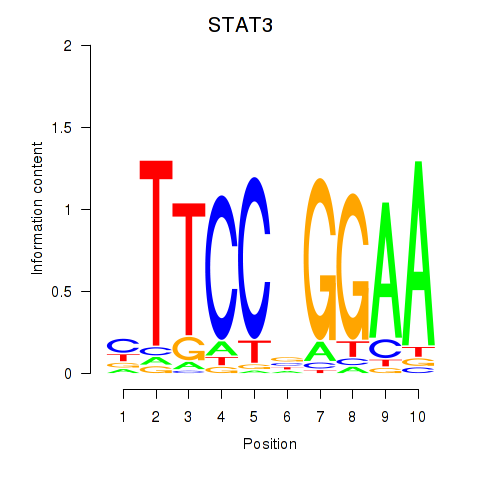
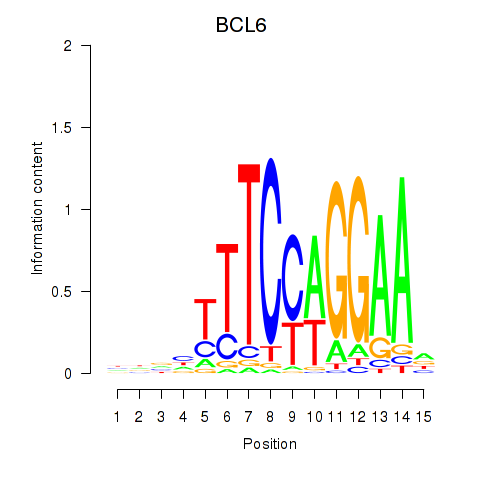
Results for STAT1_STAT3_BCL6
Z-value: 0.80
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT3 | hg19_v2_chr17_-_40540377_40540481 | -0.55 | 2.6e-01 | Click! |
| STAT1 | hg19_v2_chr2_-_191878874_191878976 | -0.50 | 3.2e-01 | Click! |
| BCL6 | hg19_v2_chr3_-_187455680_187455732 | -0.48 | 3.3e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_6625605 | 1.02 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr3_-_50649192 | 0.87 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr17_+_6347761 | 0.58 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr12_-_15082050 | 0.50 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr10_+_6625733 | 0.49 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr11_+_57531292 | 0.41 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_+_35484793 | 0.36 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr3_+_50649302 | 0.35 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr16_-_8955601 | 0.31 |
ENST00000569398.1
ENST00000568968.1 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr10_-_128110441 | 0.30 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr11_+_76493294 | 0.28 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr12_+_122064398 | 0.26 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr11_-_104817919 | 0.26 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr5_-_140013224 | 0.25 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr12_+_53443963 | 0.24 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443680 | 0.21 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_-_7244469 | 0.21 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr5_-_140013275 | 0.21 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chrX_-_154563889 | 0.21 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr3_+_122399697 | 0.20 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr16_-_67260691 | 0.20 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr17_-_41132088 | 0.19 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr5_-_146781153 | 0.19 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr9_+_70971815 | 0.17 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr2_+_87754989 | 0.17 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_+_26104104 | 0.17 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr10_+_71444655 | 0.17 |
ENST00000434931.2
|
RP11-242G20.1
|
Uncharacterized protein |
| chr22_-_31536480 | 0.17 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr6_+_46761118 | 0.17 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_+_6347729 | 0.16 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr22_-_39637135 | 0.16 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr7_+_144015218 | 0.16 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr19_+_35168567 | 0.16 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
| chr2_-_204398141 | 0.16 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr17_-_42994283 | 0.16 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr19_-_10613862 | 0.16 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr22_-_31503490 | 0.16 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr11_-_104480019 | 0.16 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr8_-_27468717 | 0.15 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr3_-_93692781 | 0.15 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr9_+_136287444 | 0.15 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr12_-_48276710 | 0.15 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr10_-_90751038 | 0.15 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr12_+_7022909 | 0.15 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr8_-_27469196 | 0.15 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr12_-_11175219 | 0.15 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr15_-_79576156 | 0.15 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr19_-_40324767 | 0.15 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr17_+_36858694 | 0.14 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr3_+_45430105 | 0.14 |
ENST00000430399.1
|
LARS2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr4_-_155511887 | 0.14 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr19_-_11688951 | 0.14 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr7_-_26578407 | 0.14 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr17_+_2264983 | 0.14 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr7_+_106415457 | 0.13 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr12_-_7245018 | 0.13 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr12_-_120687948 | 0.13 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr1_-_147632428 | 0.13 |
ENST00000599640.1
|
BX842679.1
|
Uncharacterized protein FLJ46360 |
| chr8_-_37351424 | 0.13 |
ENST00000522718.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr19_-_58864848 | 0.13 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr1_-_153521597 | 0.13 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chrX_-_153210107 | 0.13 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr6_+_131571535 | 0.13 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_67260901 | 0.13 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr12_-_49245916 | 0.13 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr15_-_41120896 | 0.12 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr15_+_57998821 | 0.12 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr11_+_844067 | 0.12 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr13_-_30160925 | 0.12 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_-_27468945 | 0.12 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr18_+_66382428 | 0.12 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr14_-_20881579 | 0.12 |
ENST00000556935.1
ENST00000262715.5 ENST00000556549.1 |
TEP1
|
telomerase-associated protein 1 |
| chr6_-_36355513 | 0.12 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr11_+_75479850 | 0.12 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr6_-_127840048 | 0.12 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr20_-_25320367 | 0.12 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr1_-_151138422 | 0.12 |
ENST00000440902.2
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr3_-_4793274 | 0.12 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr1_-_11107280 | 0.11 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr11_-_68611721 | 0.11 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chrX_-_115594160 | 0.11 |
ENST00000371894.4
|
CXorf61
|
cancer/testis antigen 83 |
| chr14_-_25479811 | 0.11 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr16_-_3030407 | 0.11 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr8_-_27468842 | 0.11 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr15_+_57998923 | 0.11 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr12_-_42877726 | 0.11 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr8_-_37351344 | 0.11 |
ENST00000520422.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr19_+_44764031 | 0.11 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr12_+_12224331 | 0.11 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr8_+_82644669 | 0.11 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr15_-_23891175 | 0.11 |
ENST00000532292.1
|
MAGEL2
|
MAGE-like 2 |
| chr19_-_10613361 | 0.10 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_+_56905024 | 0.10 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr19_+_13228917 | 0.10 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr11_+_73498973 | 0.10 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr17_-_39459103 | 0.10 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr2_+_220306238 | 0.10 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr8_-_27469383 | 0.10 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr4_-_185694872 | 0.10 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr19_+_44331555 | 0.10 |
ENST00000590950.1
|
ZNF283
|
zinc finger protein 283 |
| chr1_+_153631130 | 0.10 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr11_+_111412271 | 0.10 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr5_-_39274617 | 0.10 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr5_-_38845812 | 0.10 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr6_+_41604620 | 0.10 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr4_-_76957214 | 0.10 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr16_-_21663950 | 0.10 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr14_-_100841794 | 0.10 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr12_-_42878101 | 0.10 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr2_-_70189397 | 0.09 |
ENST00000320256.4
|
ASPRV1
|
aspartic peptidase, retroviral-like 1 |
| chr17_-_19265855 | 0.09 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr6_-_28973037 | 0.09 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr1_+_155006300 | 0.09 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr17_+_74729060 | 0.09 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr7_-_33842742 | 0.09 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr19_+_17326521 | 0.09 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_+_229440129 | 0.09 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr16_+_70332956 | 0.09 |
ENST00000288071.6
ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr14_+_76452090 | 0.09 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr5_-_119669160 | 0.09 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr14_-_45431091 | 0.09 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr5_+_61708488 | 0.09 |
ENST00000505902.1
|
IPO11
|
importin 11 |
| chr19_+_18530184 | 0.09 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr6_+_106535455 | 0.09 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr18_-_67629015 | 0.08 |
ENST00000579496.1
|
CD226
|
CD226 molecule |
| chr2_-_190044480 | 0.08 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr6_+_44194762 | 0.08 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr3_+_187871060 | 0.08 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr19_+_57106647 | 0.08 |
ENST00000328070.6
|
ZNF71
|
zinc finger protein 71 |
| chr11_-_111782484 | 0.08 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr3_+_46283863 | 0.08 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr12_-_11184006 | 0.08 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr19_-_7167989 | 0.08 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr16_-_29874211 | 0.08 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr17_+_48624450 | 0.08 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr20_+_54987305 | 0.08 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr14_-_100841670 | 0.08 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr11_-_111649074 | 0.08 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_+_77088989 | 0.08 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr6_+_292253 | 0.08 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr1_+_244227632 | 0.08 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr19_+_47835404 | 0.08 |
ENST00000595464.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr22_+_45725524 | 0.08 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr14_-_81408063 | 0.08 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr19_+_49128209 | 0.08 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr17_-_26697304 | 0.08 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr11_+_66025091 | 0.08 |
ENST00000526758.1
ENST00000440228.2 |
KLC2
|
kinesin light chain 2 |
| chr1_+_153004800 | 0.08 |
ENST00000392661.3
|
SPRR1B
|
small proline-rich protein 1B |
| chr19_-_44809121 | 0.07 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chrX_+_107288280 | 0.07 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr18_-_53253323 | 0.07 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr14_+_32963433 | 0.07 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_-_144995074 | 0.07 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_+_17935383 | 0.07 |
ENST00000524465.1
ENST00000381254.2 ENST00000532257.1 |
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr6_+_31730773 | 0.07 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr6_+_108977520 | 0.07 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr3_+_62304648 | 0.07 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr7_+_76026832 | 0.07 |
ENST00000336517.4
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor) |
| chr12_-_58165870 | 0.07 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr1_-_151138323 | 0.07 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr1_+_165864800 | 0.07 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr6_-_39197226 | 0.07 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chrX_+_129473859 | 0.07 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_27760247 | 0.07 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr16_-_66519747 | 0.07 |
ENST00000544589.1
|
AC132186.1
|
CDNA FLJ27243 fis, clone SYN08134; Uncharacterized protein |
| chr7_-_100881041 | 0.07 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr1_+_114472992 | 0.07 |
ENST00000514621.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_-_28113217 | 0.07 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr11_-_6462210 | 0.07 |
ENST00000265983.3
|
HPX
|
hemopexin |
| chr1_+_182584314 | 0.07 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr15_+_84322827 | 0.07 |
ENST00000286744.5
ENST00000567476.1 |
ADAMTSL3
|
ADAMTS-like 3 |
| chr11_+_46740730 | 0.07 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr11_-_9781068 | 0.07 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr19_+_10196981 | 0.07 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_7618413 | 0.07 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_-_44405623 | 0.07 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chrX_-_62571187 | 0.07 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr6_+_63921399 | 0.07 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr19_+_44331444 | 0.07 |
ENST00000324461.7
|
ZNF283
|
zinc finger protein 283 |
| chr19_+_44598492 | 0.06 |
ENST00000589680.1
|
ZNF224
|
zinc finger protein 224 |
| chr4_+_86396265 | 0.06 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_62323702 | 0.06 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr16_+_67563250 | 0.06 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr5_+_140501581 | 0.06 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr1_+_15668240 | 0.06 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr1_+_6511651 | 0.06 |
ENST00000434576.1
|
ESPN
|
espin |
| chr2_+_101591314 | 0.06 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr17_-_38210644 | 0.06 |
ENST00000394128.2
ENST00000394127.2 ENST00000356271.3 ENST00000535071.2 ENST00000580885.1 ENST00000543759.2 ENST00000537674.2 ENST00000580517.1 ENST00000578161.1 |
MED24
|
mediator complex subunit 24 |
| chr16_-_89785777 | 0.06 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chrX_-_2882296 | 0.06 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr11_+_65657875 | 0.06 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr16_+_67261008 | 0.06 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr9_+_125796806 | 0.06 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr17_+_76311791 | 0.06 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr3_-_148939835 | 0.06 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr19_-_1155118 | 0.06 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr2_-_197675000 | 0.06 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr1_-_161207986 | 0.06 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_-_27135591 | 0.06 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr19_+_18530146 | 0.06 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.5 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.1 | 0.2 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0090677 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:2000360 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) negative regulation of fertilization(GO:0060467) acrosomal vesicle exocytosis(GO:0060478) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.0 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:1902024 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.0 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.0 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.0 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0015227 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.0 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |