Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
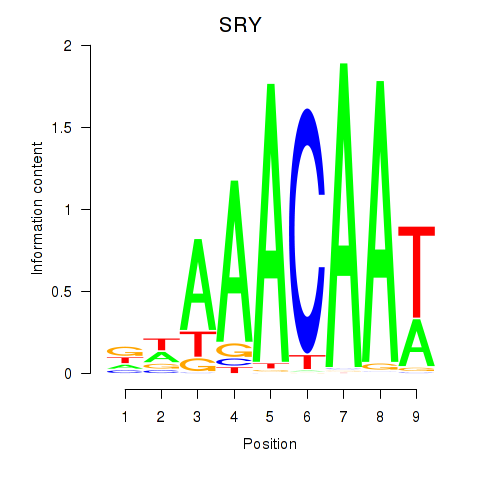
Results for SRY
Z-value: 0.64
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
Activity profile of SRY motif
Sorted Z-values of SRY motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_178865887 | 0.48 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr8_-_71157595 | 0.39 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr14_+_38065052 | 0.37 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chrX_+_107288280 | 0.35 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_162101247 | 0.33 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr10_+_70847852 | 0.31 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr4_-_111120334 | 0.30 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr17_-_57229155 | 0.30 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_+_21666633 | 0.29 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr8_-_95449155 | 0.28 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr3_-_28390120 | 0.25 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr7_+_134551583 | 0.24 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr3_-_28389922 | 0.23 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr12_+_78359999 | 0.23 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr12_-_102591604 | 0.23 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr2_-_55237484 | 0.23 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr3_-_28390298 | 0.23 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr14_-_89960395 | 0.22 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr4_+_24661479 | 0.22 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr11_-_46848393 | 0.22 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr14_-_55878538 | 0.22 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chrX_+_78003204 | 0.22 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr2_-_207078086 | 0.20 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr14_-_38036271 | 0.19 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr17_-_46671323 | 0.19 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr5_+_110073853 | 0.19 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr11_-_72852320 | 0.19 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr6_-_56716686 | 0.18 |
ENST00000520645.1
|
DST
|
dystonin |
| chr18_+_20715416 | 0.18 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr3_-_28390415 | 0.18 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr3_+_112930387 | 0.17 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr8_-_42358742 | 0.17 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_+_1832902 | 0.17 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr10_-_77161533 | 0.16 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr12_-_76817036 | 0.16 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_+_137673200 | 0.16 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr17_-_70053866 | 0.16 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr14_-_58893876 | 0.16 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_53019211 | 0.15 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr4_-_76598326 | 0.15 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_+_109204909 | 0.15 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_-_49504449 | 0.15 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr15_+_91416092 | 0.15 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr4_-_105416039 | 0.15 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr13_+_110958124 | 0.14 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr4_-_103266626 | 0.14 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr5_-_111091948 | 0.14 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chrX_+_123095860 | 0.13 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chrX_+_123095890 | 0.13 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr3_+_178866199 | 0.13 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr1_+_25664408 | 0.13 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr1_-_40367530 | 0.13 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_+_84630574 | 0.13 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_37126765 | 0.13 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr18_-_13915530 | 0.13 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr20_-_62582475 | 0.13 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr4_-_76598296 | 0.12 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr15_+_49715449 | 0.12 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr3_+_112929850 | 0.12 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr17_-_49124230 | 0.12 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chrX_+_9880412 | 0.12 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr10_-_77161650 | 0.12 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr10_-_32667660 | 0.12 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr2_-_207078154 | 0.12 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr3_-_192635943 | 0.12 |
ENST00000392452.2
|
MB21D2
|
Mab-21 domain containing 2 |
| chr11_+_36317830 | 0.12 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr4_+_41614909 | 0.12 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chrX_+_9880590 | 0.12 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr4_-_111120132 | 0.12 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr18_+_11851383 | 0.11 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr2_-_192711968 | 0.11 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr6_+_53794780 | 0.11 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr7_-_78400598 | 0.11 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_+_93645314 | 0.11 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr11_+_28724129 | 0.11 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr9_-_128246769 | 0.10 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr2_-_157198860 | 0.10 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr14_+_58894404 | 0.10 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr11_-_10879572 | 0.10 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr3_+_69812701 | 0.10 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_152146385 | 0.10 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr14_+_58894103 | 0.09 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr3_-_57233966 | 0.09 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr1_+_84630053 | 0.09 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_109089573 | 0.09 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr3_-_123339418 | 0.09 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr6_-_134639042 | 0.09 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_116381918 | 0.09 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr8_-_145688231 | 0.09 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr22_+_19710468 | 0.09 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr3_-_134092561 | 0.09 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr16_+_53469525 | 0.09 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr18_-_64271316 | 0.09 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr17_-_73511584 | 0.09 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr17_+_27573875 | 0.08 |
ENST00000225387.3
|
CRYBA1
|
crystallin, beta A1 |
| chr14_-_36990061 | 0.08 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr18_+_46065570 | 0.08 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr3_-_28390581 | 0.08 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr14_-_23288930 | 0.08 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr18_-_53019208 | 0.08 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr5_+_133859996 | 0.08 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr4_+_76871883 | 0.08 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr11_+_128563652 | 0.08 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_-_70994612 | 0.07 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr21_-_19191703 | 0.07 |
ENST00000284881.4
ENST00000400559.3 ENST00000400558.3 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr5_-_137674000 | 0.07 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr17_+_79953310 | 0.07 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr2_+_181988620 | 0.07 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr8_-_145060593 | 0.07 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr1_-_115053781 | 0.07 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr2_+_27665289 | 0.07 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr4_-_65275162 | 0.07 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr5_-_131132614 | 0.07 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr17_-_46703826 | 0.07 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr17_-_79212825 | 0.07 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr19_+_13134772 | 0.07 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_100316041 | 0.07 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chrX_+_123095546 | 0.07 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr3_-_141868293 | 0.07 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_58652523 | 0.07 |
ENST00000489857.1
ENST00000358781.2 |
FAM3D
|
family with sequence similarity 3, member D |
| chr4_-_68749745 | 0.07 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr2_+_181988560 | 0.07 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr10_+_11207485 | 0.07 |
ENST00000537122.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr8_-_116673894 | 0.07 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_-_13570533 | 0.06 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr15_-_70390213 | 0.06 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr15_+_49715293 | 0.06 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr7_+_7811992 | 0.06 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr13_-_74708372 | 0.06 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr1_-_93645818 | 0.06 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr2_+_30569506 | 0.06 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr6_-_37665751 | 0.06 |
ENST00000297153.7
ENST00000434837.3 |
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr9_+_132099158 | 0.06 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr20_-_61002584 | 0.06 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr20_-_22566089 | 0.06 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr3_+_141596371 | 0.06 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr14_+_58894141 | 0.06 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr10_+_11207088 | 0.06 |
ENST00000608830.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_-_73511504 | 0.06 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr14_+_58894706 | 0.06 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr4_+_41614720 | 0.06 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_-_62149433 | 0.06 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr16_-_4852915 | 0.06 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr18_+_18943554 | 0.06 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr4_-_76598544 | 0.05 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_+_33722080 | 0.05 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr1_+_164528866 | 0.05 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr3_+_142342228 | 0.05 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr1_+_84630352 | 0.05 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_91580732 | 0.05 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr20_-_52612705 | 0.05 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_+_118955500 | 0.05 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr4_-_109090106 | 0.05 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr1_+_235490659 | 0.05 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_-_204436344 | 0.05 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_+_91581011 | 0.05 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr15_-_55563072 | 0.05 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_+_55712915 | 0.05 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_-_46272462 | 0.05 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr19_+_13135790 | 0.05 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr17_-_79212884 | 0.05 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr14_-_89883412 | 0.05 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr8_+_24151620 | 0.05 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr3_+_57741957 | 0.05 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr20_-_43150601 | 0.04 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr5_+_137673945 | 0.04 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr3_-_47205066 | 0.04 |
ENST00000412450.1
|
SETD2
|
SET domain containing 2 |
| chr12_-_62586543 | 0.04 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr2_-_190927447 | 0.04 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr3_+_189507432 | 0.04 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr3_-_11610255 | 0.04 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr10_-_105845674 | 0.04 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr17_+_37809333 | 0.04 |
ENST00000443521.1
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr4_-_140477910 | 0.04 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr2_-_160473114 | 0.04 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr7_+_30185496 | 0.04 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr21_-_32931290 | 0.04 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr3_-_135913298 | 0.04 |
ENST00000434835.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr1_-_40367668 | 0.04 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr10_+_115312825 | 0.04 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr1_+_84630645 | 0.04 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_-_161085291 | 0.04 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr2_+_208423891 | 0.04 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr5_+_140248518 | 0.04 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr14_-_61124977 | 0.04 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr16_+_53920795 | 0.04 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr2_+_181845843 | 0.04 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr8_-_13134045 | 0.04 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr1_+_164529004 | 0.04 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr5_-_131132658 | 0.04 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr1_+_33116765 | 0.03 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chr15_+_90544532 | 0.03 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr5_+_147774275 | 0.03 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr16_-_69390209 | 0.03 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr2_+_27665232 | 0.03 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr5_+_86563636 | 0.03 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr20_+_32150140 | 0.03 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_+_86947296 | 0.03 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr15_+_59397275 | 0.03 |
ENST00000288207.2
|
CCNB2
|
cyclin B2 |
| chr3_+_189507523 | 0.03 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.0 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0045013 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.0 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |