Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
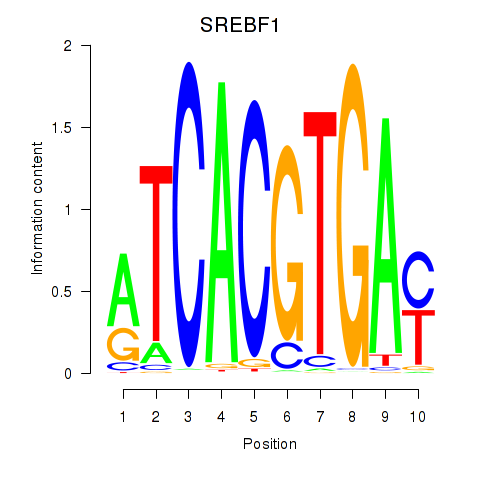
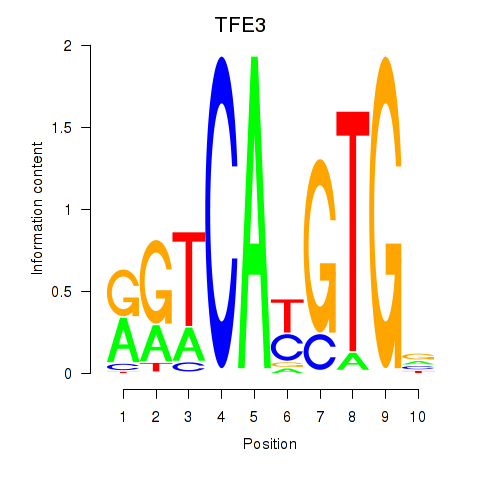
Results for SREBF1_TFE3
Z-value: 0.65
Transcription factors associated with SREBF1_TFE3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF1
|
ENSG00000072310.12 | sterol regulatory element binding transcription factor 1 |
|
TFE3
|
ENSG00000068323.12 | transcription factor binding to IGHM enhancer 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SREBF1 | hg19_v2_chr17_-_17740287_17740316 | 0.51 | 3.0e-01 | Click! |
| TFE3 | hg19_v2_chrX_-_48901012_48901050 | -0.35 | 4.9e-01 | Click! |
Activity profile of SREBF1_TFE3 motif
Sorted Z-values of SREBF1_TFE3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_72668805 | 0.88 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr11_-_1785139 | 0.71 |
ENST00000236671.2
|
CTSD
|
cathepsin D |
| chr16_-_5083917 | 0.61 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr17_+_42422629 | 0.58 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr17_+_78075361 | 0.52 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr19_-_36545649 | 0.50 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr17_+_78075498 | 0.50 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr16_+_28986134 | 0.49 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr17_-_7137582 | 0.47 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr7_+_4815238 | 0.46 |
ENST00000348624.4
ENST00000401897.1 |
AP5Z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr19_+_10812108 | 0.44 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr4_+_159131596 | 0.43 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr17_+_42422637 | 0.42 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr19_-_10764509 | 0.42 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr17_+_42422662 | 0.40 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chrX_-_102565932 | 0.40 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr17_+_1627834 | 0.40 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr20_-_44519839 | 0.40 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr17_+_6915730 | 0.39 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr19_+_40873617 | 0.38 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr4_+_76439095 | 0.38 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr16_+_2570431 | 0.38 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr19_+_40854559 | 0.38 |
ENST00000598962.1
ENST00000409419.1 ENST00000409587.1 ENST00000602131.1 ENST00000409735.4 ENST00000600948.1 ENST00000356508.5 ENST00000596682.1 ENST00000594908.1 |
PLD3
|
phospholipase D family, member 3 |
| chr22_-_36903069 | 0.37 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_+_6915798 | 0.36 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr16_+_28986085 | 0.35 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr8_-_11660077 | 0.35 |
ENST00000533405.1
|
RP11-297N6.4
|
Uncharacterized protein |
| chr16_+_4421841 | 0.34 |
ENST00000304735.3
|
VASN
|
vasorin |
| chr16_-_5083589 | 0.33 |
ENST00000563578.1
ENST00000562346.2 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr19_-_5680891 | 0.33 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr1_-_154193091 | 0.33 |
ENST00000362076.4
ENST00000350592.3 ENST00000368516.1 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr17_-_7137857 | 0.33 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr3_-_49395705 | 0.32 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr11_+_67159416 | 0.32 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr6_-_33385870 | 0.31 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr3_-_49395892 | 0.31 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chr19_-_40854281 | 0.30 |
ENST00000392035.2
|
C19orf47
|
chromosome 19 open reading frame 47 |
| chr16_-_1525016 | 0.30 |
ENST00000262318.8
ENST00000448525.1 |
CLCN7
|
chloride channel, voltage-sensitive 7 |
| chr19_-_1237990 | 0.30 |
ENST00000382477.2
ENST00000215376.6 ENST00000590083.1 |
C19orf26
|
chromosome 19 open reading frame 26 |
| chr16_+_2570340 | 0.29 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_-_67514982 | 0.29 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr17_+_3539744 | 0.28 |
ENST00000046640.3
ENST00000381870.3 |
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr9_+_131084846 | 0.28 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr1_-_156265438 | 0.27 |
ENST00000362007.1
|
C1orf85
|
chromosome 1 open reading frame 85 |
| chr16_-_28503357 | 0.27 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr13_+_113951607 | 0.27 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr17_+_73975292 | 0.26 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr16_-_28503080 | 0.26 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr20_-_36156264 | 0.26 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr15_+_44084040 | 0.26 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr17_-_76124812 | 0.25 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr17_+_78075324 | 0.25 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chr11_-_1606513 | 0.25 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr5_-_180242576 | 0.25 |
ENST00000514438.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr7_+_99746514 | 0.25 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr9_-_90589402 | 0.25 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr20_+_44519948 | 0.25 |
ENST00000354880.5
ENST00000191018.5 |
CTSA
|
cathepsin A |
| chr6_-_33385854 | 0.25 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr13_+_113951532 | 0.24 |
ENST00000332556.4
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr7_+_100271446 | 0.24 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_+_242673994 | 0.24 |
ENST00000321264.4
ENST00000537090.1 ENST00000403782.1 ENST00000342518.6 |
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase |
| chr3_+_50654821 | 0.24 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr10_+_69644404 | 0.24 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr20_+_44520009 | 0.24 |
ENST00000607482.1
ENST00000372459.2 |
CTSA
|
cathepsin A |
| chr17_-_65992544 | 0.23 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr16_-_28503327 | 0.23 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr3_+_51428704 | 0.23 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr17_+_6915902 | 0.23 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr1_+_153004800 | 0.23 |
ENST00000392661.3
|
SPRR1B
|
small proline-rich protein 1B |
| chr10_+_99258625 | 0.22 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr19_-_40854417 | 0.22 |
ENST00000582006.1
ENST00000582783.1 |
C19orf47
|
chromosome 19 open reading frame 47 |
| chr7_+_100271355 | 0.22 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_-_49016418 | 0.22 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr22_-_20850128 | 0.22 |
ENST00000328879.4
|
KLHL22
|
kelch-like family member 22 |
| chr7_-_27196267 | 0.21 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr19_+_7587555 | 0.21 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr19_+_7587491 | 0.21 |
ENST00000264079.6
|
MCOLN1
|
mucolipin 1 |
| chr19_+_10764937 | 0.21 |
ENST00000449870.1
ENST00000318511.3 ENST00000420083.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr9_-_131709858 | 0.21 |
ENST00000372586.3
|
DOLK
|
dolichol kinase |
| chr6_-_44225231 | 0.21 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr1_+_10093188 | 0.20 |
ENST00000377153.1
|
UBE4B
|
ubiquitination factor E4B |
| chrX_-_102565858 | 0.20 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr19_+_5681153 | 0.20 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr2_-_47572207 | 0.20 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr7_-_27135591 | 0.20 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr1_-_27682962 | 0.20 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr5_-_1112141 | 0.20 |
ENST00000264930.5
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporter), member 7 |
| chr5_+_149865377 | 0.20 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_-_220042825 | 0.19 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr19_-_48018203 | 0.19 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr8_-_145688231 | 0.19 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr4_-_113627966 | 0.19 |
ENST00000505632.1
|
RP11-148B6.2
|
RP11-148B6.2 |
| chr17_+_3539998 | 0.19 |
ENST00000452111.1
ENST00000574776.1 ENST00000441220.2 ENST00000414524.2 |
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr16_-_425205 | 0.19 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr9_-_90589586 | 0.19 |
ENST00000325303.8
ENST00000375883.3 |
CDK20
|
cyclin-dependent kinase 20 |
| chr12_+_112451222 | 0.19 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr16_+_5083950 | 0.19 |
ENST00000588623.1
|
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr22_+_38203898 | 0.19 |
ENST00000323205.6
ENST00000248924.6 ENST00000445195.1 |
GCAT
|
glycine C-acetyltransferase |
| chr11_-_71814422 | 0.19 |
ENST00000278671.5
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr19_+_1275917 | 0.19 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr12_-_90103077 | 0.18 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_240323439 | 0.18 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr5_-_110074603 | 0.18 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr22_-_36903101 | 0.18 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr1_+_152957707 | 0.18 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr11_-_110968081 | 0.18 |
ENST00000603154.1
|
RP11-89C3.4
|
RP11-89C3.4 |
| chr19_+_797392 | 0.18 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr16_+_88923494 | 0.17 |
ENST00000567895.1
ENST00000301021.3 ENST00000565504.1 ENST00000567312.1 ENST00000568583.1 ENST00000561840.1 |
TRAPPC2L
|
trafficking protein particle complex 2-like |
| chr22_+_21996549 | 0.17 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr2_+_120187465 | 0.17 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr6_-_163834852 | 0.17 |
ENST00000604200.1
|
CAHM
|
colon adenocarcinoma hypermethylated (non-protein coding) |
| chr15_-_76352069 | 0.17 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_-_76124711 | 0.17 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr17_-_79791118 | 0.17 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr11_-_71814276 | 0.17 |
ENST00000538404.1
ENST00000535107.1 ENST00000545249.1 |
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr12_+_51633061 | 0.17 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr17_+_79935418 | 0.17 |
ENST00000306729.7
ENST00000306739.4 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr17_-_73851285 | 0.17 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr19_-_3985455 | 0.17 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr19_-_36499521 | 0.17 |
ENST00000397428.3
ENST00000503121.1 ENST00000340477.5 ENST00000324444.3 ENST00000490730.1 |
SYNE4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr3_-_19988462 | 0.17 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr7_+_99070527 | 0.17 |
ENST00000379724.3
|
ZNF789
|
zinc finger protein 789 |
| chr14_-_45431091 | 0.17 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr5_-_154230130 | 0.16 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr1_+_44440575 | 0.16 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr1_-_155211017 | 0.16 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr14_+_24458093 | 0.16 |
ENST00000558753.1
ENST00000537912.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr19_-_13227534 | 0.16 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr4_-_100242549 | 0.16 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr20_+_18118486 | 0.16 |
ENST00000432901.3
|
PET117
|
PET117 homolog (S. cerevisiae) |
| chr19_+_6464243 | 0.16 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr16_+_2098003 | 0.16 |
ENST00000439673.2
ENST00000350773.4 |
TSC2
|
tuberous sclerosis 2 |
| chr16_+_2097970 | 0.16 |
ENST00000382538.6
ENST00000401874.2 ENST00000353929.4 |
TSC2
|
tuberous sclerosis 2 |
| chr14_-_94443065 | 0.15 |
ENST00000555287.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr7_+_100464760 | 0.15 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr7_+_102073966 | 0.15 |
ENST00000495936.1
ENST00000356387.2 ENST00000478730.2 ENST00000468241.1 ENST00000403646.3 |
ORAI2
|
ORAI calcium release-activated calcium modulator 2 |
| chr19_-_5720248 | 0.15 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr5_-_176730676 | 0.15 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr12_-_109027643 | 0.15 |
ENST00000388962.3
ENST00000550948.1 |
SELPLG
|
selectin P ligand |
| chr19_-_8386238 | 0.15 |
ENST00000301457.2
|
NDUFA7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr2_+_220042933 | 0.15 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr7_-_150777920 | 0.15 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr9_+_131084815 | 0.15 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr19_-_5680499 | 0.15 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr4_-_87028478 | 0.15 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_-_175964366 | 0.15 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr19_-_15236470 | 0.14 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr11_+_66360665 | 0.14 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase |
| chr19_-_40324767 | 0.14 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr9_-_34665983 | 0.14 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr14_-_106068065 | 0.14 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr17_+_37356528 | 0.14 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr12_-_108154705 | 0.14 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr6_+_87865262 | 0.14 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr22_-_31536480 | 0.14 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr1_-_154909329 | 0.14 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr19_+_6464502 | 0.14 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr17_+_37356555 | 0.14 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr7_+_74379083 | 0.14 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr20_-_2821271 | 0.14 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr19_-_36545128 | 0.14 |
ENST00000538849.1
|
THAP8
|
THAP domain containing 8 |
| chr8_-_71157595 | 0.14 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr6_-_33385902 | 0.14 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_101360374 | 0.14 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr19_-_12833361 | 0.14 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr5_-_159739483 | 0.14 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr19_-_13227463 | 0.14 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr13_+_36920569 | 0.13 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr8_-_145331153 | 0.13 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr20_+_2821340 | 0.13 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr9_-_91793675 | 0.13 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr19_-_5340730 | 0.13 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr5_+_140514782 | 0.13 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chrX_+_10126488 | 0.13 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr19_-_3500635 | 0.13 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr21_-_38445470 | 0.13 |
ENST00000399098.1
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_-_125473600 | 0.13 |
ENST00000308736.2
|
DHX37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chrX_+_153657009 | 0.13 |
ENST00000449556.1
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1 |
| chr12_+_51632666 | 0.13 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr19_+_40854363 | 0.13 |
ENST00000599685.1
ENST00000392032.2 |
PLD3
|
phospholipase D family, member 3 |
| chr16_-_75150665 | 0.12 |
ENST00000300051.4
ENST00000450168.2 |
LDHD
|
lactate dehydrogenase D |
| chr1_+_92545862 | 0.12 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr19_-_18632861 | 0.12 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr16_-_30134441 | 0.12 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr3_-_122512619 | 0.12 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr12_+_117176090 | 0.12 |
ENST00000257575.4
ENST00000407967.3 ENST00000392549.2 |
RNFT2
|
ring finger protein, transmembrane 2 |
| chr3_+_127317066 | 0.12 |
ENST00000265056.7
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr14_+_105941118 | 0.12 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr11_-_63376013 | 0.12 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr19_+_12848299 | 0.12 |
ENST00000357332.3
|
ASNA1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr6_+_138188351 | 0.12 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_572543 | 0.12 |
ENST00000333511.3
ENST00000573216.1 ENST00000353555.4 |
BSG
|
basigin (Ok blood group) |
| chr1_-_155211065 | 0.12 |
ENST00000427500.3
|
GBA
|
glucosidase, beta, acid |
| chr3_+_158519654 | 0.12 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr22_-_39268308 | 0.12 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr6_-_84937314 | 0.12 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr6_+_139349903 | 0.12 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr20_-_2821756 | 0.12 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr1_+_11796177 | 0.12 |
ENST00000400895.2
ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP
|
angiotensin II receptor-associated protein |
| chr3_-_20227720 | 0.12 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF1_TFE3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.2 | 0.8 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.8 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.6 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.3 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.2 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.1 | 0.2 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.3 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.2 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.9 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.3 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 | 0.6 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.3 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:2000340 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.0 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0030210 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:2000851 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:1902164 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0035350 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 3.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.0 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.4 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 0.3 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.0 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.5 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0002060 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |