Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
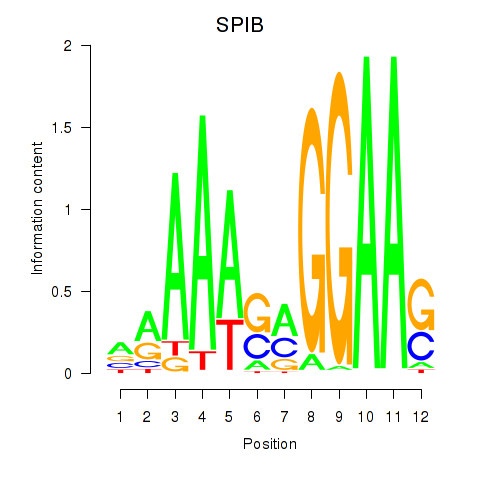
Results for SPIB
Z-value: 1.02
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | -0.18 | 7.3e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_25205155 | 0.81 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr3_+_121774202 | 0.58 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr12_+_25205446 | 0.57 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr14_+_38065052 | 0.55 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr12_-_15114191 | 0.55 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_179851611 | 0.53 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr8_-_101733794 | 0.52 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_+_27062272 | 0.49 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_+_121796697 | 0.45 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr8_+_125860939 | 0.45 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr10_-_98031265 | 0.44 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr17_-_39123144 | 0.44 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr11_+_27062860 | 0.43 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_25205568 | 0.43 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_25205666 | 0.42 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr10_-_98031310 | 0.42 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr12_-_15114492 | 0.40 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_+_25205628 | 0.40 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr8_+_74903580 | 0.40 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr15_-_83224682 | 0.39 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr7_+_23636992 | 0.38 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr8_-_131028782 | 0.38 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr17_-_5138099 | 0.36 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr13_+_108921977 | 0.33 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr11_+_27062502 | 0.33 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_-_35618364 | 0.32 |
ENST00000378431.1
ENST00000378430.3 ENST00000259633.4 |
CD72
|
CD72 molecule |
| chrX_-_11129229 | 0.32 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr15_-_33360085 | 0.30 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chr3_-_180397256 | 0.30 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr20_-_20033052 | 0.28 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr12_-_15114658 | 0.28 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_112016414 | 0.27 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_+_120839005 | 0.27 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr19_+_42381337 | 0.27 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr5_-_39270725 | 0.27 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr12_-_51785083 | 0.27 |
ENST00000603563.1
|
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr16_+_70695570 | 0.26 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr6_-_56708459 | 0.26 |
ENST00000370788.2
|
DST
|
dystonin |
| chr11_-_64570624 | 0.26 |
ENST00000439069.1
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr3_-_176914191 | 0.25 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr8_-_74206133 | 0.25 |
ENST00000352983.2
|
RPL7
|
ribosomal protein L7 |
| chr1_+_164528616 | 0.25 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_86525299 | 0.25 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_20227720 | 0.25 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chrX_+_105937068 | 0.25 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_206138884 | 0.24 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr3_+_184530173 | 0.24 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr16_+_30960375 | 0.24 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr3_+_184529948 | 0.24 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr12_+_7060508 | 0.24 |
ENST00000541698.1
ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_-_20227619 | 0.24 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr19_-_54726850 | 0.23 |
ENST00000245620.9
ENST00000346401.6 ENST00000424807.1 ENST00000445347.1 |
LILRB3
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr5_-_142783365 | 0.22 |
ENST00000508760.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_184529929 | 0.22 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_-_176914238 | 0.22 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr13_+_53602894 | 0.22 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr11_-_1606513 | 0.21 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr20_-_46415341 | 0.21 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr8_-_101734170 | 0.21 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_26644441 | 0.21 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr14_+_32546145 | 0.21 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_120839412 | 0.21 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr7_+_23637118 | 0.21 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_+_36190853 | 0.20 |
ENST00000433436.2
ENST00000538225.1 ENST00000540080.1 |
CLTA
|
clathrin, light chain A |
| chr3_-_143567262 | 0.20 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr15_+_86098670 | 0.20 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr3_-_9291063 | 0.20 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr1_+_110158726 | 0.20 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr6_-_56707943 | 0.20 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr9_+_36190905 | 0.20 |
ENST00000345519.5
ENST00000470744.1 ENST00000242285.6 ENST00000466396.1 ENST00000396603.2 |
CLTA
|
clathrin, light chain A |
| chr10_+_104613980 | 0.20 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr1_-_31230650 | 0.20 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr12_+_32638897 | 0.19 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_-_51471362 | 0.19 |
ENST00000376853.4
ENST00000424910.2 |
KLK6
|
kallikrein-related peptidase 6 |
| chr8_-_101734308 | 0.19 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_-_158757895 | 0.19 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr9_+_34653861 | 0.19 |
ENST00000556792.1
ENST00000318041.9 ENST00000378817.4 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr12_+_7060414 | 0.18 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_30721968 | 0.18 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr2_+_233925064 | 0.18 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr20_-_46415297 | 0.18 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr6_-_110012380 | 0.18 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr5_-_140013275 | 0.17 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr18_-_52989525 | 0.17 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr22_-_37880543 | 0.17 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_104068312 | 0.17 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr2_+_102758271 | 0.17 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr4_-_140005443 | 0.17 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr3_+_23847432 | 0.17 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr2_+_127413677 | 0.17 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr10_+_7830125 | 0.17 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chrX_-_47489244 | 0.17 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr11_+_45168182 | 0.16 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr2_-_74405929 | 0.16 |
ENST00000396049.4
|
MOB1A
|
MOB kinase activator 1A |
| chr8_-_121457608 | 0.16 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr2_-_8723918 | 0.16 |
ENST00000454224.1
|
AC011747.4
|
AC011747.4 |
| chr2_-_231084820 | 0.16 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr5_+_96079240 | 0.16 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr3_+_46283863 | 0.16 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chrX_+_123094672 | 0.16 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr1_+_167599532 | 0.16 |
ENST00000537350.1
|
RCSD1
|
RCSD domain containing 1 |
| chr5_+_95997769 | 0.16 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr10_+_32856764 | 0.16 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr14_+_75536335 | 0.16 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr20_-_57607347 | 0.16 |
ENST00000395663.1
ENST00000395659.1 ENST00000243997.3 |
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr19_+_48828582 | 0.16 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr11_-_64889252 | 0.16 |
ENST00000525297.1
ENST00000529259.1 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chrX_+_105936982 | 0.15 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr13_-_36050819 | 0.15 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr19_+_35940486 | 0.15 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr13_-_95364389 | 0.15 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr12_-_15082050 | 0.15 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr1_-_45956822 | 0.15 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr10_+_7830092 | 0.15 |
ENST00000356708.7
|
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr15_+_50716576 | 0.15 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chrX_+_152990302 | 0.15 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr14_+_75536280 | 0.15 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr11_+_1860832 | 0.15 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_-_64751227 | 0.15 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr17_-_27467418 | 0.15 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr3_-_11685345 | 0.15 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr4_+_95129061 | 0.15 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_130339710 | 0.15 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr11_-_61124711 | 0.15 |
ENST00000536915.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr19_-_13213662 | 0.15 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr2_+_127413704 | 0.15 |
ENST00000409836.3
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr3_-_179169330 | 0.15 |
ENST00000232564.3
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr5_+_159848854 | 0.15 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr2_-_152830441 | 0.14 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr15_+_83209620 | 0.14 |
ENST00000568285.1
|
RP11-379H8.1
|
Uncharacterized protein |
| chr1_+_222886694 | 0.14 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr19_-_17375541 | 0.14 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr7_-_135662056 | 0.14 |
ENST00000393085.3
ENST00000435723.1 |
MTPN
|
myotrophin |
| chr4_+_110834033 | 0.14 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr10_-_62149433 | 0.14 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr11_+_1861399 | 0.14 |
ENST00000381905.3
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr14_+_85996507 | 0.13 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_+_24616588 | 0.13 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr2_-_174830430 | 0.13 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr11_-_61124776 | 0.13 |
ENST00000542361.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr15_+_51973550 | 0.13 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr4_+_159727222 | 0.13 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr14_+_63671105 | 0.13 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr1_-_145039949 | 0.13 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr14_+_85996471 | 0.13 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_-_87281224 | 0.13 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_+_3875757 | 0.13 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr5_-_142783175 | 0.13 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_-_49430886 | 0.12 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr5_-_142782862 | 0.12 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_+_159727272 | 0.12 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_+_196440692 | 0.12 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr10_-_33623310 | 0.12 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr1_-_43282906 | 0.12 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr8_-_74205851 | 0.12 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr3_-_28390581 | 0.12 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr9_-_21995249 | 0.12 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr9_+_214842 | 0.12 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr1_-_222886526 | 0.12 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr5_+_64859066 | 0.12 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr8_+_98656693 | 0.12 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr9_-_21995300 | 0.12 |
ENST00000498628.2
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr2_-_152118352 | 0.11 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr5_+_102455968 | 0.11 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_-_45956800 | 0.11 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr1_+_161551101 | 0.11 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr2_-_153573887 | 0.11 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_-_140005341 | 0.11 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr3_+_28390637 | 0.11 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr3_+_29322437 | 0.11 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr1_+_46713404 | 0.11 |
ENST00000371975.4
ENST00000469835.1 |
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr10_-_65028817 | 0.11 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr5_-_124084493 | 0.11 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr17_+_28443819 | 0.11 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr19_+_7413835 | 0.11 |
ENST00000576789.1
|
CTB-133G6.1
|
CTB-133G6.1 |
| chrX_+_48398053 | 0.11 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr17_+_28443799 | 0.11 |
ENST00000584423.1
ENST00000247026.5 |
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr15_-_65809991 | 0.11 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr3_-_178984759 | 0.11 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr8_-_121457332 | 0.11 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr7_-_117512264 | 0.11 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr18_-_47018769 | 0.11 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr14_+_96968707 | 0.10 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr11_-_8680383 | 0.10 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr14_+_96968802 | 0.10 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chrX_-_30595959 | 0.10 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr15_-_33360342 | 0.10 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr10_-_52008313 | 0.10 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr2_+_138722028 | 0.10 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr14_-_75536182 | 0.10 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr14_-_98444369 | 0.10 |
ENST00000554822.1
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr19_-_16770915 | 0.10 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr8_-_131028869 | 0.10 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr3_+_4535155 | 0.10 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr17_+_34948228 | 0.10 |
ENST00000251312.5
ENST00000590554.1 |
DHRS11
|
dehydrogenase/reductase (SDR family) member 11 |
| chr18_-_32924372 | 0.10 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr1_-_220445757 | 0.10 |
ENST00000358951.2
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr8_-_95449155 | 0.10 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_+_167298281 | 0.10 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr20_-_35274548 | 0.10 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.2 | 1.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.3 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.5 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0002877 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.1 | 0.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.3 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.6 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0043317 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.2 | GO:1905205 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.5 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.0 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:1902378 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) sensory neuron axon guidance(GO:0097374) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 2.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.0 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0061485 | memory T cell proliferation(GO:0061485) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0071723 | peptidoglycan receptor activity(GO:0016019) lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |