Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
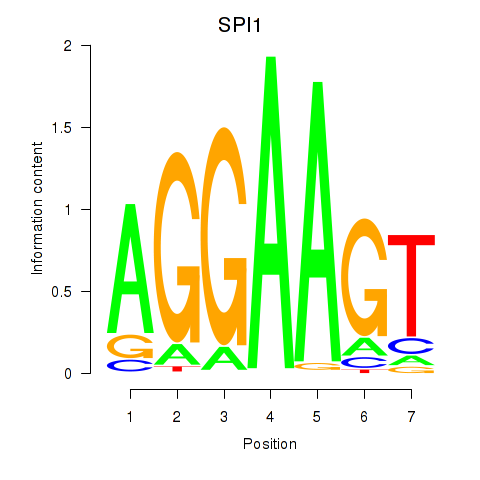
Results for SPI1
Z-value: 0.92
Transcription factors associated with SPI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPI1
|
ENSG00000066336.7 | Spi-1 proto-oncogene |
Activity profile of SPI1 motif
Sorted Z-values of SPI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_38171681 | 2.30 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr17_+_38171614 | 1.63 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr17_+_67590125 | 1.60 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr2_+_228678550 | 1.31 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr5_+_159895275 | 1.25 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr15_-_80263506 | 1.17 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_+_31543334 | 0.90 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr6_+_31895467 | 0.66 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_-_114429997 | 0.66 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr11_+_18154059 | 0.65 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr6_+_26156551 | 0.65 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_+_31895480 | 0.60 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr5_-_150460914 | 0.54 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_102826434 | 0.52 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr6_+_144980954 | 0.50 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr1_+_153330322 | 0.48 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr22_-_30642728 | 0.48 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr1_-_183560011 | 0.45 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr5_+_35856951 | 0.45 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr8_-_42065075 | 0.44 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr11_+_102188272 | 0.41 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr6_-_11382478 | 0.41 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr8_-_42065187 | 0.40 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr10_-_75532373 | 0.39 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr17_+_45908974 | 0.38 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr14_-_69445793 | 0.38 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr16_+_4845379 | 0.38 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr8_-_82395461 | 0.37 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr11_+_102188224 | 0.37 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr7_+_105172612 | 0.37 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr8_+_61592073 | 0.36 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr11_-_102714534 | 0.36 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr2_+_138721850 | 0.36 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr2_-_153573965 | 0.35 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_-_235098935 | 0.35 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr17_-_38859996 | 0.34 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr1_+_152881014 | 0.34 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr3_+_119013185 | 0.34 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr6_-_34524049 | 0.33 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr14_-_69445968 | 0.33 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr3_+_142838091 | 0.32 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr17_+_44668387 | 0.32 |
ENST00000576040.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr9_+_108424738 | 0.32 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr17_-_1463095 | 0.31 |
ENST00000575895.1
ENST00000573056.1 |
PITPNA
|
phosphatidylinositol transfer protein, alpha |
| chr11_-_3862059 | 0.31 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr1_-_183559693 | 0.30 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr17_-_40134339 | 0.30 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr6_-_41908428 | 0.30 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr1_+_152943122 | 0.29 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr19_+_6772710 | 0.29 |
ENST00000304076.2
ENST00000602142.1 ENST00000596764.1 |
VAV1
|
vav 1 guanine nucleotide exchange factor |
| chr17_+_6918064 | 0.28 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr18_-_72264805 | 0.28 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr12_+_96588143 | 0.28 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr6_+_139349903 | 0.28 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr17_-_48277552 | 0.28 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr14_-_69446034 | 0.28 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr1_-_159880159 | 0.28 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr6_-_32811771 | 0.27 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_4710288 | 0.27 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr8_+_124194875 | 0.27 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr17_+_6918093 | 0.26 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr6_-_34524093 | 0.26 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr8_-_25281747 | 0.26 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr3_+_9774164 | 0.26 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr11_-_64815333 | 0.25 |
ENST00000533842.1
ENST00000532802.1 ENST00000530139.1 ENST00000526516.1 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr4_+_103422499 | 0.25 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr6_-_32160622 | 0.25 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr2_+_37571717 | 0.25 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr17_+_7461580 | 0.24 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_+_12391685 | 0.24 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr4_-_40516560 | 0.24 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr15_+_90544532 | 0.24 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr9_+_134001455 | 0.24 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr2_-_96811170 | 0.23 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr16_-_30125177 | 0.23 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_152214098 | 0.23 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_5569588 | 0.23 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr9_-_139268068 | 0.23 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr4_-_77819002 | 0.23 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr20_+_31823792 | 0.23 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr19_+_15218180 | 0.23 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr6_-_43595039 | 0.23 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr11_-_6502534 | 0.22 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr6_+_30951487 | 0.22 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr1_-_52520828 | 0.22 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr18_+_22040593 | 0.22 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr12_-_92539614 | 0.22 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr9_-_134145880 | 0.21 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr19_-_12792585 | 0.21 |
ENST00000351660.5
|
DHPS
|
deoxyhypusine synthase |
| chr16_+_2732476 | 0.21 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr17_+_41561317 | 0.21 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chrX_-_55020511 | 0.21 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr11_-_58343319 | 0.21 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr3_-_121379739 | 0.21 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_-_156786634 | 0.21 |
ENST00000392306.2
ENST00000368199.3 |
SH2D2A
|
SH2 domain containing 2A |
| chr1_-_161039647 | 0.21 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr7_-_27135591 | 0.21 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr20_+_3767547 | 0.20 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr17_-_1029052 | 0.20 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr10_-_75351088 | 0.20 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr20_+_2633269 | 0.20 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr3_+_47324424 | 0.20 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr15_-_64386120 | 0.20 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr6_+_30689350 | 0.20 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr10_-_75676400 | 0.20 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr7_+_114055052 | 0.20 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr15_-_40398812 | 0.20 |
ENST00000561360.1
|
BMF
|
Bcl2 modifying factor |
| chr11_-_6502580 | 0.20 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr21_+_17909594 | 0.20 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_982198 | 0.19 |
ENST00000571945.1
ENST00000536794.2 |
ABR
|
active BCR-related |
| chr6_+_33589161 | 0.19 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr4_+_103423055 | 0.19 |
ENST00000505458.1
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr6_-_161695042 | 0.19 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr19_-_6481776 | 0.19 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr19_+_45251804 | 0.19 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chrX_+_154114635 | 0.19 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr6_+_30689401 | 0.19 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr1_-_150738261 | 0.19 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr8_-_28965684 | 0.19 |
ENST00000523130.1
|
KIF13B
|
kinesin family member 13B |
| chrX_-_19689106 | 0.19 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_+_219262948 | 0.19 |
ENST00000443891.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr7_+_106505696 | 0.19 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr6_-_161695074 | 0.19 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_-_161039753 | 0.18 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr8_+_99076509 | 0.18 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr17_+_43299241 | 0.18 |
ENST00000328118.3
|
FMNL1
|
formin-like 1 |
| chr17_+_6918354 | 0.18 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr2_+_198669365 | 0.18 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr5_-_40835303 | 0.18 |
ENST00000509877.1
ENST00000508493.1 ENST00000274242.5 |
RPL37
|
ribosomal protein L37 |
| chr4_+_81256871 | 0.18 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr1_-_235098861 | 0.18 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr7_+_106505912 | 0.18 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr15_+_75074915 | 0.18 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr16_-_28506840 | 0.18 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr1_-_159924006 | 0.18 |
ENST00000368092.3
ENST00000368093.3 |
SLAMF9
|
SLAM family member 9 |
| chr15_+_90744533 | 0.18 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_-_155947951 | 0.18 |
ENST00000313695.7
ENST00000497907.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_43833628 | 0.18 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr7_+_157129660 | 0.18 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr6_-_167040731 | 0.18 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr16_+_69373323 | 0.17 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr8_+_56792355 | 0.17 |
ENST00000519728.1
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr15_+_75074410 | 0.17 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr20_+_5731083 | 0.17 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr1_-_205912577 | 0.17 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr17_-_6917755 | 0.17 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr2_+_169659121 | 0.17 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr12_+_96588368 | 0.17 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr7_-_45957011 | 0.17 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr17_-_7760457 | 0.17 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr8_+_24151553 | 0.17 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr17_-_18907463 | 0.17 |
ENST00000585154.2
ENST00000345041.4 |
FAM83G
|
family with sequence similarity 83, member G |
| chr17_-_40913029 | 0.17 |
ENST00000592195.1
ENST00000592670.1 ENST00000587694.1 ENST00000591082.1 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr11_-_8739383 | 0.17 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr7_-_45956856 | 0.17 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr22_+_38004723 | 0.17 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr8_+_56792377 | 0.17 |
ENST00000520220.2
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr16_-_279405 | 0.17 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr1_+_179851893 | 0.17 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr4_+_103422471 | 0.17 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr4_+_8201091 | 0.17 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr16_-_66968055 | 0.17 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr11_-_47270341 | 0.17 |
ENST00000529444.1
ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr3_-_16555150 | 0.17 |
ENST00000334133.4
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr15_+_77713299 | 0.17 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr2_+_68962014 | 0.16 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr15_-_64385981 | 0.16 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr17_+_7462103 | 0.16 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_-_98031155 | 0.16 |
ENST00000495266.1
|
BLNK
|
B-cell linker |
| chr7_-_45018686 | 0.16 |
ENST00000258787.7
|
MYO1G
|
myosin IG |
| chr12_-_123201337 | 0.16 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr6_-_159466136 | 0.16 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr11_-_102401469 | 0.16 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr6_-_154751629 | 0.16 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr5_-_43412418 | 0.16 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr1_-_212588157 | 0.16 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr16_-_66968265 | 0.16 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr11_+_131781290 | 0.16 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr1_-_207095324 | 0.16 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chrX_-_100662881 | 0.16 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr4_+_2626988 | 0.16 |
ENST00000509050.1
|
FAM193A
|
family with sequence similarity 193, member A |
| chr11_+_64794875 | 0.16 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr21_+_33671160 | 0.16 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr6_-_155635583 | 0.16 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr1_-_161039456 | 0.16 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr17_+_7255208 | 0.16 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr14_+_102276192 | 0.16 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_+_50388126 | 0.16 |
ENST00000425346.1
ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2
XXcos-LUCA11.5
|
cytochrome b561 family, member D2 Uncharacterized protein |
| chr5_-_131826457 | 0.16 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr3_-_161089289 | 0.16 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr17_-_40913275 | 0.16 |
ENST00000589716.1
ENST00000360166.3 |
RAMP2-AS1
|
RAMP2 antisense RNA 1 |
| chr1_+_41174988 | 0.15 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr5_+_139505520 | 0.15 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr11_+_120081475 | 0.15 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr8_+_124194752 | 0.15 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr3_-_192445289 | 0.15 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr6_-_33290580 | 0.15 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr17_+_18218587 | 0.15 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr16_-_67693846 | 0.15 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr22_-_31328881 | 0.15 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 0.9 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 | 3.9 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.6 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.5 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.3 | GO:0070666 | regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 0.5 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.6 | GO:0060481 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) peptidyl-glutamine modification(GO:0018199) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.2 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.9 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.5 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.1 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.1 | 0.2 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.2 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 1.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0098751 | megakaryocyte development(GO:0035855) bone cell development(GO:0098751) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0033590 | response to cobalamin(GO:0033590) response to erythropoietin(GO:0036017) cellular response to erythropoietin(GO:0036018) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.2 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.1 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0061354 | chemoattraction of serotonergic neuron axon(GO:0036517) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.0 | 0.0 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.9 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.5 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.5 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.2 | 0.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.6 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 2.8 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 6.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |