Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
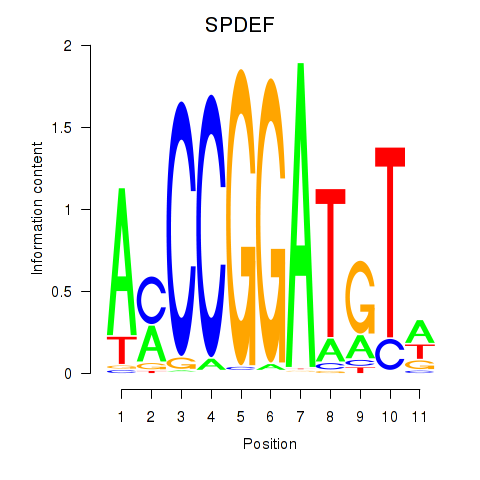
Results for SPDEF
Z-value: 0.96
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSG00000124664.6 | SAM pointed domain containing ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPDEF | hg19_v2_chr6_-_34524049_34524091 | -0.97 | 1.2e-03 | Click! |
Activity profile of SPDEF motif
Sorted Z-values of SPDEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_105212297 | 0.74 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr5_+_158690089 | 0.70 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr4_+_141445333 | 0.54 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr7_-_77427676 | 0.49 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr4_-_153700864 | 0.47 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr14_-_31926623 | 0.46 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr8_-_100025238 | 0.44 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr17_-_56065540 | 0.44 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr18_-_19180681 | 0.43 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr14_-_67826538 | 0.41 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr4_+_48833312 | 0.41 |
ENST00000508293.1
ENST00000513391.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr15_-_65282274 | 0.36 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_+_84629976 | 0.35 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr11_-_71751715 | 0.34 |
ENST00000535947.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_63815628 | 0.34 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr4_+_166128735 | 0.34 |
ENST00000226725.6
|
KLHL2
|
kelch-like family member 2 |
| chr1_+_67395922 | 0.34 |
ENST00000401042.3
ENST00000355356.3 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr15_-_65282232 | 0.33 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_+_84630053 | 0.33 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_-_119806085 | 0.33 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr1_+_193028552 | 0.33 |
ENST00000400968.2
ENST00000432079.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr7_-_38948774 | 0.33 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr14_-_31926595 | 0.32 |
ENST00000547378.1
|
RP11-176H8.1
|
Uncharacterized protein |
| chr14_+_60716276 | 0.32 |
ENST00000528241.2
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr17_+_65714018 | 0.32 |
ENST00000581106.1
ENST00000535137.1 |
NOL11
|
nucleolar protein 11 |
| chr9_-_72374848 | 0.32 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr17_+_65713925 | 0.31 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr14_-_64010006 | 0.31 |
ENST00000555899.1
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr15_-_59949667 | 0.31 |
ENST00000396061.1
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr12_+_102513950 | 0.31 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr3_+_32023232 | 0.31 |
ENST00000360311.4
|
ZNF860
|
zinc finger protein 860 |
| chr12_-_57914275 | 0.30 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr7_-_91875109 | 0.30 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr7_+_77428066 | 0.30 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr15_-_64648273 | 0.29 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr8_-_124665190 | 0.29 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr5_-_114961858 | 0.29 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr4_+_48833183 | 0.28 |
ENST00000503016.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr10_-_14996070 | 0.28 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr12_+_102514019 | 0.27 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr12_+_29302119 | 0.27 |
ENST00000536681.3
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr7_+_77428149 | 0.27 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr8_+_15397732 | 0.27 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr3_-_139108463 | 0.26 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr2_+_198380289 | 0.26 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr6_+_134274354 | 0.26 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr1_+_235492300 | 0.25 |
ENST00000476121.1
ENST00000497327.1 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr2_+_162016827 | 0.25 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_80987617 | 0.25 |
ENST00000258884.4
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C |
| chr2_+_63816126 | 0.25 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr19_-_45004556 | 0.25 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr8_-_77912431 | 0.25 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr14_-_31926701 | 0.24 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr12_+_29302023 | 0.24 |
ENST00000551451.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr4_+_39640787 | 0.24 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr7_-_91875358 | 0.24 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr1_-_193028426 | 0.24 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr13_+_25670268 | 0.24 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr1_+_170501270 | 0.23 |
ENST00000367763.3
ENST00000367762.1 |
GORAB
|
golgin, RAB6-interacting |
| chr4_-_83812157 | 0.23 |
ENST00000513323.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr14_+_60716159 | 0.23 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr4_+_144106063 | 0.23 |
ENST00000510377.1
|
USP38
|
ubiquitin specific peptidase 38 |
| chr14_-_69263043 | 0.23 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_193028621 | 0.22 |
ENST00000367455.4
ENST00000367454.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr4_+_48833119 | 0.22 |
ENST00000444354.2
ENST00000509963.1 ENST00000509246.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr2_-_54014127 | 0.22 |
ENST00000394717.2
|
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chrX_-_131352152 | 0.22 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr5_+_110074685 | 0.22 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr8_-_56986768 | 0.22 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr18_+_48556470 | 0.21 |
ENST00000589076.1
ENST00000590061.1 ENST00000591914.1 ENST00000342988.3 |
SMAD4
|
SMAD family member 4 |
| chr8_-_130951940 | 0.21 |
ENST00000522250.1
ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr20_-_34287220 | 0.21 |
ENST00000306750.3
|
NFS1
|
NFS1 cysteine desulfurase |
| chr6_-_143832793 | 0.21 |
ENST00000438118.2
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr19_+_38880695 | 0.21 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_+_49961990 | 0.21 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr14_-_64010046 | 0.20 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr2_+_27805880 | 0.20 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr9_+_36190853 | 0.20 |
ENST00000433436.2
ENST00000538225.1 ENST00000540080.1 |
CLTA
|
clathrin, light chain A |
| chr18_+_11851383 | 0.20 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr2_+_101618691 | 0.20 |
ENST00000409000.1
ENST00000409028.4 ENST00000409320.3 ENST00000409733.1 ENST00000409650.1 ENST00000409038.1 |
RPL31
|
ribosomal protein L31 |
| chr3_-_133380731 | 0.20 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr10_-_15902449 | 0.19 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr16_-_25122785 | 0.19 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr7_-_135662056 | 0.19 |
ENST00000393085.3
ENST00000435723.1 |
MTPN
|
myotrophin |
| chr19_-_53696587 | 0.19 |
ENST00000396424.3
ENST00000600412.1 |
ZNF665
|
zinc finger protein 665 |
| chr9_+_36190905 | 0.19 |
ENST00000345519.5
ENST00000470744.1 ENST00000242285.6 ENST00000466396.1 ENST00000396603.2 |
CLTA
|
clathrin, light chain A |
| chr4_+_48833015 | 0.19 |
ENST00000509122.1
ENST00000509664.1 ENST00000505922.2 ENST00000514981.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr12_-_64616019 | 0.19 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr1_+_235491714 | 0.19 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr17_-_33905521 | 0.19 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr2_+_217363793 | 0.19 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr15_-_59949693 | 0.19 |
ENST00000396063.1
ENST00000396064.3 ENST00000484743.1 ENST00000559706.1 ENST00000396060.2 |
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr7_+_93551011 | 0.19 |
ENST00000248564.5
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr2_+_162016804 | 0.18 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_118122996 | 0.18 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr1_+_193028717 | 0.18 |
ENST00000415442.2
ENST00000506303.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr17_-_7108436 | 0.18 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr1_-_169337176 | 0.18 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr14_+_60715928 | 0.18 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr8_-_101964738 | 0.18 |
ENST00000523938.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_+_48833160 | 0.18 |
ENST00000506801.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr14_+_67826709 | 0.17 |
ENST00000256383.4
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr2_+_63816087 | 0.17 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr3_-_37217736 | 0.17 |
ENST00000438374.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_-_54014055 | 0.17 |
ENST00000263634.3
ENST00000406687.1 |
GPR75-ASB3
|
GPR75-ASB3 readthrough |
| chr1_+_22351977 | 0.17 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr5_+_76326187 | 0.17 |
ENST00000312916.7
ENST00000506806.1 |
AGGF1
|
angiogenic factor with G patch and FHA domains 1 |
| chr3_+_197464046 | 0.17 |
ENST00000428738.1
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr2_+_162016916 | 0.16 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_-_69864993 | 0.16 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr2_+_37311588 | 0.16 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr2_+_39103103 | 0.16 |
ENST00000340556.6
ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2
|
MORN repeat containing 2 |
| chr20_+_60962143 | 0.16 |
ENST00000343986.4
|
RPS21
|
ribosomal protein S21 |
| chr4_+_48833234 | 0.16 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr11_-_119993734 | 0.16 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr14_+_21566980 | 0.16 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr1_+_84630574 | 0.16 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_99613195 | 0.15 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr12_+_106751436 | 0.15 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr4_-_44728559 | 0.15 |
ENST00000509756.1
ENST00000507917.1 ENST00000295448.3 |
GNPDA2
|
glucosamine-6-phosphate deaminase 2 |
| chr12_+_50135351 | 0.15 |
ENST00000549445.1
ENST00000550951.1 ENST00000549385.1 ENST00000548713.1 ENST00000548201.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr8_-_56987061 | 0.15 |
ENST00000009589.3
ENST00000524349.1 ENST00000519606.1 ENST00000519807.1 ENST00000521262.1 ENST00000520627.1 |
RPS20
|
ribosomal protein S20 |
| chr2_+_48667898 | 0.15 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr15_-_65809581 | 0.15 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr21_-_46221684 | 0.15 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr4_+_144106080 | 0.15 |
ENST00000307017.4
|
USP38
|
ubiquitin specific peptidase 38 |
| chr18_+_21032781 | 0.15 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr17_-_56082455 | 0.15 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr3_-_156272924 | 0.15 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr4_+_41992489 | 0.14 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr14_+_24701870 | 0.14 |
ENST00000561035.1
ENST00000559409.1 ENST00000558865.1 ENST00000558279.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr7_+_114562172 | 0.14 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_29322437 | 0.14 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr6_-_150039249 | 0.14 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr8_+_8860314 | 0.14 |
ENST00000250263.7
ENST00000519292.1 |
ERI1
|
exoribonuclease 1 |
| chr2_+_27805971 | 0.14 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chrX_-_122866874 | 0.14 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr2_+_65454926 | 0.13 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr12_-_69080590 | 0.13 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr8_-_101964832 | 0.13 |
ENST00000523131.1
ENST00000418997.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_+_159593418 | 0.13 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr6_-_150039170 | 0.13 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr2_+_47596634 | 0.13 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr7_+_99613212 | 0.13 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_-_235324772 | 0.13 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr12_+_69080734 | 0.13 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr1_+_65886244 | 0.13 |
ENST00000344610.8
|
LEPR
|
leptin receptor |
| chr14_-_24615523 | 0.13 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr18_+_21033239 | 0.13 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr1_+_84630645 | 0.13 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_+_19321281 | 0.12 |
ENST00000261537.6
|
MIB1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr2_+_65454863 | 0.12 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr3_-_37218023 | 0.12 |
ENST00000416425.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_+_50135327 | 0.12 |
ENST00000549966.1
ENST00000547832.1 ENST00000547187.1 ENST00000548894.1 ENST00000546914.1 ENST00000552699.1 ENST00000267115.5 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr5_+_93954039 | 0.12 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr11_-_61197406 | 0.12 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr14_+_24701819 | 0.11 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr12_-_120663792 | 0.11 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr2_+_37311645 | 0.11 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr15_-_44092295 | 0.11 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chr10_+_5726764 | 0.11 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr1_+_24829384 | 0.11 |
ENST00000374395.4
ENST00000436717.2 |
RCAN3
|
RCAN family member 3 |
| chr18_-_55288973 | 0.11 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr17_-_47755436 | 0.10 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr3_-_69101461 | 0.10 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr16_-_70557430 | 0.10 |
ENST00000393612.4
ENST00000564653.1 ENST00000323786.5 |
COG4
|
component of oligomeric golgi complex 4 |
| chr12_+_132434439 | 0.10 |
ENST00000333577.4
ENST00000389561.2 ENST00000389562.2 ENST00000332482.4 ENST00000330386.6 |
EP400
|
E1A binding protein p400 |
| chr19_-_8579030 | 0.10 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr5_-_114961673 | 0.10 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr6_+_7541808 | 0.10 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr1_+_213224572 | 0.10 |
ENST00000543470.1
ENST00000366960.3 ENST00000366959.3 ENST00000543354.1 |
RPS6KC1
|
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
| chr1_+_169337172 | 0.10 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr11_+_36616355 | 0.09 |
ENST00000532470.2
|
C11orf74
|
chromosome 11 open reading frame 74 |
| chr22_-_41215291 | 0.09 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr11_+_61197508 | 0.09 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr21_-_36421626 | 0.09 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_-_193028632 | 0.09 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr6_-_116381918 | 0.09 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr10_-_14996017 | 0.09 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr11_-_46142948 | 0.09 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr8_-_101734170 | 0.09 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_43124087 | 0.09 |
ENST00000304979.3
ENST00000372550.1 ENST00000440068.1 |
PPIH
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr4_-_159592996 | 0.09 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr19_-_56632592 | 0.09 |
ENST00000587279.1
ENST00000270459.3 |
ZNF787
|
zinc finger protein 787 |
| chr5_+_31532373 | 0.09 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr11_+_57480046 | 0.09 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr20_+_49126881 | 0.09 |
ENST00000371621.3
ENST00000541713.1 |
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr11_-_61129306 | 0.08 |
ENST00000544118.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr14_+_24702073 | 0.08 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr6_-_143832820 | 0.08 |
ENST00000002165.6
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr12_+_118814185 | 0.08 |
ENST00000543473.1
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr16_-_2318111 | 0.08 |
ENST00000301730.8
ENST00000564822.1 |
RNPS1
|
RNA binding protein S1, serine-rich domain |
| chr3_-_69101413 | 0.08 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chr15_-_55700457 | 0.08 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr1_+_169337412 | 0.08 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr8_-_101965104 | 0.08 |
ENST00000437293.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_-_57479673 | 0.08 |
ENST00000337672.2
ENST00000431606.2 |
MED19
|
mediator complex subunit 19 |
| chr2_+_187350883 | 0.08 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_-_94706705 | 0.08 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr12_-_102513843 | 0.08 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPDEF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 1.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.5 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.2 | GO:0035262 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) gonad morphogenesis(GO:0035262) |
| 0.1 | 0.3 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.2 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.3 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.3 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.3 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.6 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.2 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.2 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.3 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.7 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.4 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.1 | 0.3 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.2 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |