Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
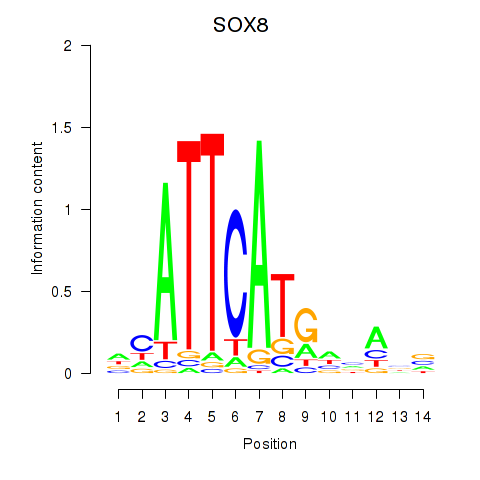
Results for SOX8
Z-value: 0.75
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | -0.13 | 8.1e-01 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153029980 | 0.69 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr1_+_153330322 | 0.65 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr11_+_18154059 | 0.61 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr12_+_113354341 | 0.60 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_+_13677795 | 0.56 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr14_+_95078714 | 0.56 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr1_+_81106951 | 0.51 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_+_152956549 | 0.48 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr8_+_125283924 | 0.44 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr22_-_30642728 | 0.42 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr10_-_4285835 | 0.34 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr2_-_25451065 | 0.34 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr14_-_21562671 | 0.33 |
ENST00000554923.1
|
ZNF219
|
zinc finger protein 219 |
| chr2_-_36779411 | 0.33 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr12_-_96793142 | 0.32 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr15_+_91445448 | 0.31 |
ENST00000558290.1
ENST00000558853.1 ENST00000559999.1 |
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr17_-_43339474 | 0.28 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr16_+_82068830 | 0.28 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_82068873 | 0.26 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr8_-_11873043 | 0.26 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr22_-_50708781 | 0.26 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr16_-_28503357 | 0.25 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr3_-_54962100 | 0.23 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr4_+_22999152 | 0.22 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr3_-_193272741 | 0.22 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr2_-_214017151 | 0.22 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_-_200992827 | 0.22 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr20_-_5426380 | 0.22 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr15_+_45938079 | 0.21 |
ENST00000561493.1
|
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr7_+_6714599 | 0.21 |
ENST00000328239.7
ENST00000542006.1 |
AC073343.1
|
Uncharacterized protein |
| chrX_+_18725758 | 0.21 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr4_+_75174204 | 0.21 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr10_-_75676400 | 0.20 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr22_+_45714301 | 0.20 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr2_+_219646462 | 0.20 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr10_-_4285923 | 0.20 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr6_-_131211534 | 0.20 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr18_-_73967160 | 0.20 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr5_+_89770696 | 0.20 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr19_-_21950332 | 0.19 |
ENST00000598026.1
|
ZNF100
|
zinc finger protein 100 |
| chr7_+_6713376 | 0.19 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr3_-_100558953 | 0.19 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr17_+_39421591 | 0.19 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr10_-_101825151 | 0.19 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr11_-_9482010 | 0.19 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr11_+_12308447 | 0.18 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr12_-_50101165 | 0.18 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chr17_-_27230035 | 0.18 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr6_-_133079022 | 0.18 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr5_+_118788138 | 0.18 |
ENST00000256216.6
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr12_-_12714025 | 0.18 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr19_+_1269324 | 0.17 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr19_-_6333614 | 0.17 |
ENST00000301452.4
|
ACER1
|
alkaline ceramidase 1 |
| chr15_+_45926919 | 0.17 |
ENST00000561735.1
ENST00000260324.7 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr5_+_89770664 | 0.17 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr10_+_9012087 | 0.17 |
ENST00000456526.1
|
RP11-428L9.2
|
RP11-428L9.2 |
| chr17_-_43339453 | 0.17 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr17_+_2240916 | 0.17 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr17_-_41132410 | 0.17 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr4_+_9446156 | 0.16 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr11_-_125366089 | 0.16 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_+_135596106 | 0.16 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr20_-_1472029 | 0.16 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr2_-_228497888 | 0.15 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr14_+_75745477 | 0.15 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr11_+_57435219 | 0.14 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr12_-_50101003 | 0.14 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr6_+_80129989 | 0.14 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr16_-_65106110 | 0.14 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr4_+_89299994 | 0.14 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_+_49711391 | 0.14 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr7_+_18548878 | 0.13 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr14_-_68159152 | 0.13 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr12_+_24857397 | 0.13 |
ENST00000536131.1
|
RP11-625L16.1
|
RP11-625L16.1 |
| chr20_+_48807351 | 0.13 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr3_+_38347427 | 0.13 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr2_+_204801471 | 0.13 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr5_-_68339648 | 0.13 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr10_-_74114714 | 0.13 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr4_-_137020391 | 0.13 |
ENST00000569694.1
|
RP11-775H9.3
|
RP11-775H9.3 |
| chr12_-_49581152 | 0.13 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr14_+_32963433 | 0.13 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr9_-_128246769 | 0.13 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr7_+_18548924 | 0.12 |
ENST00000524023.1
|
HDAC9
|
histone deacetylase 9 |
| chr4_-_70518941 | 0.12 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr16_+_57769635 | 0.12 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr6_+_42018614 | 0.12 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr3_+_98216448 | 0.12 |
ENST00000427338.1
|
OR5K2
|
olfactory receptor, family 5, subfamily K, member 2 |
| chr12_-_123201337 | 0.12 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr9_-_130661916 | 0.12 |
ENST00000373142.1
ENST00000373146.1 ENST00000373144.3 |
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr1_-_203274418 | 0.12 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr17_+_7387677 | 0.12 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr1_+_38512799 | 0.12 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr15_+_81299370 | 0.12 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr10_-_81932771 | 0.12 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr16_+_87636474 | 0.11 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr7_-_92855762 | 0.11 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr18_+_71815743 | 0.11 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr2_+_33359473 | 0.11 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_159237472 | 0.11 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr17_+_22022437 | 0.11 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chrX_+_49091920 | 0.11 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr2_-_3584430 | 0.11 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr5_-_160279207 | 0.11 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr1_-_183538319 | 0.11 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_66820058 | 0.11 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_+_57392684 | 0.11 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr12_+_48876275 | 0.11 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr20_-_47804894 | 0.11 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr2_-_170550877 | 0.11 |
ENST00000447353.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr20_+_32399093 | 0.11 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr16_+_30076052 | 0.11 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_166326157 | 0.11 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr16_+_30387141 | 0.11 |
ENST00000566955.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_+_156680153 | 0.10 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr6_-_76072719 | 0.10 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_+_173116225 | 0.10 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_-_140544386 | 0.10 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr19_+_39903185 | 0.10 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr16_+_30075783 | 0.10 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr9_+_99690592 | 0.10 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr12_-_11548496 | 0.10 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr16_+_22308717 | 0.10 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr5_+_140227048 | 0.10 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr12_-_11508520 | 0.10 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr3_-_141719195 | 0.10 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_102615416 | 0.10 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr6_+_31021225 | 0.10 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr9_-_113761720 | 0.10 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr11_+_10477733 | 0.10 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr2_+_97203082 | 0.10 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr2_+_113816215 | 0.10 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr8_-_91095099 | 0.09 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr19_+_19144384 | 0.09 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr19_+_45681997 | 0.09 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr1_+_161475208 | 0.09 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr12_+_1099675 | 0.09 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr16_+_30075595 | 0.09 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr18_+_32455201 | 0.09 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr8_+_7752151 | 0.09 |
ENST00000302247.2
|
DEFB4A
|
defensin, beta 4A |
| chr16_+_82068585 | 0.09 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr10_-_128110441 | 0.09 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr13_-_47012325 | 0.09 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr18_-_5396271 | 0.09 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr12_-_123187890 | 0.09 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr12_+_93130311 | 0.09 |
ENST00000344636.3
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr2_-_37501692 | 0.09 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr5_-_146781153 | 0.09 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr16_+_31271274 | 0.08 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr3_-_52090461 | 0.08 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr16_+_87985029 | 0.08 |
ENST00000439677.1
ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP
|
BTG3 associated nuclear protein |
| chr6_+_37012607 | 0.08 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr6_-_138539627 | 0.08 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr8_+_87111059 | 0.08 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr6_-_134638767 | 0.08 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_+_110769258 | 0.08 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr9_+_27524283 | 0.08 |
ENST00000276943.2
|
IFNK
|
interferon, kappa |
| chr5_+_156887027 | 0.08 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr17_-_34207295 | 0.08 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr7_+_17414360 | 0.08 |
ENST00000419463.1
|
AC019117.1
|
AC019117.1 |
| chr5_+_140186647 | 0.08 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr5_-_133510456 | 0.08 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr11_+_43702322 | 0.08 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr18_-_3845293 | 0.08 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_151344172 | 0.08 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr6_-_52668605 | 0.08 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr4_-_10686475 | 0.07 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr3_-_49851313 | 0.07 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr3_-_49459878 | 0.07 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr5_-_132948216 | 0.07 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr18_-_33702078 | 0.07 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr6_-_32160622 | 0.07 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr10_-_131762105 | 0.07 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr2_-_163695128 | 0.07 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr14_-_21562648 | 0.07 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr3_+_173302222 | 0.07 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr11_-_128457446 | 0.07 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr18_-_29264467 | 0.07 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr14_+_35591928 | 0.07 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr7_-_14026123 | 0.07 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr3_+_154958715 | 0.07 |
ENST00000462531.1
ENST00000490497.1 |
RP11-451G4.1
|
RP11-451G4.1 |
| chr1_+_182584314 | 0.07 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr17_-_66951474 | 0.07 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr10_+_7745232 | 0.06 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr18_-_3845321 | 0.06 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_120124258 | 0.06 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr15_+_25101698 | 0.06 |
ENST00000400097.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr15_+_80733570 | 0.06 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_-_1779663 | 0.06 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr19_-_42806842 | 0.06 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr12_+_58176525 | 0.06 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr4_-_144940477 | 0.06 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr2_-_70944855 | 0.06 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr2_+_170550944 | 0.06 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr15_+_62853562 | 0.06 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr8_+_30496078 | 0.06 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr4_+_156680143 | 0.06 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr10_-_21186144 | 0.06 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr15_+_40674920 | 0.06 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr12_-_76879852 | 0.06 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.3 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0061571 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:1902080 | negative regulation of calcium-transporting ATPase activity(GO:1901895) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0052364 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 0.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.1 | GO:0052813 | phosphatidylinositol 3-kinase activity(GO:0035004) phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.0 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |