Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
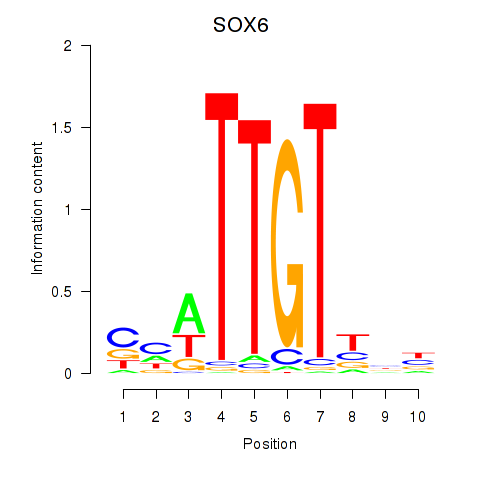
Results for SOX6
Z-value: 0.54
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16419067_16419108 | 0.29 | 5.8e-01 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_166702601 | 0.60 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr10_-_75351088 | 0.36 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr12_-_58145889 | 0.30 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr6_+_13272904 | 0.29 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr16_-_3068171 | 0.28 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr13_+_51913819 | 0.21 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr6_-_127840048 | 0.21 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr12_-_11508520 | 0.20 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr1_+_152943122 | 0.19 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr2_-_114647327 | 0.19 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr4_-_186732892 | 0.19 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_76037081 | 0.18 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr20_-_52645231 | 0.18 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_+_95611536 | 0.16 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_231989808 | 0.16 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr12_-_11548496 | 0.15 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr3_-_69402828 | 0.15 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr6_+_32936942 | 0.15 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr12_-_8815299 | 0.15 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr12_+_95611569 | 0.15 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_11905413 | 0.14 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr8_-_11660077 | 0.14 |
ENST00000533405.1
|
RP11-297N6.4
|
Uncharacterized protein |
| chr17_+_1666108 | 0.13 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_17571722 | 0.13 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr2_-_208031542 | 0.12 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_85430356 | 0.12 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr20_-_1317555 | 0.12 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr22_+_37959647 | 0.12 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr4_-_68620053 | 0.12 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr6_-_31782813 | 0.11 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr6_-_167040693 | 0.11 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr12_+_95611516 | 0.11 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr20_-_39317868 | 0.11 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr4_-_186732048 | 0.11 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_105416039 | 0.11 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr13_-_41593425 | 0.11 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr4_-_186732241 | 0.11 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr22_+_39052632 | 0.11 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr17_+_72667239 | 0.10 |
ENST00000402449.4
|
RAB37
|
RAB37, member RAS oncogene family |
| chr1_+_14075903 | 0.10 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr3_-_114790179 | 0.10 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_+_4773205 | 0.10 |
ENST00000440139.1
|
CDYL
|
chromodomain protein, Y-like |
| chr18_+_7946839 | 0.10 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_+_54243917 | 0.10 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr3_-_52719888 | 0.10 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr12_+_124997766 | 0.10 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr11_-_75236867 | 0.09 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr17_-_56065484 | 0.09 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chrX_+_107068959 | 0.09 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr12_-_58146048 | 0.09 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr4_+_26322987 | 0.09 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_+_15528332 | 0.09 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr3_-_167813672 | 0.09 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr3_-_167813132 | 0.09 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr3_+_186649133 | 0.09 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_+_89631647 | 0.09 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chrX_+_80457442 | 0.09 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr4_+_54243798 | 0.08 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr11_-_10830463 | 0.08 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr11_+_46402482 | 0.08 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr10_+_22605304 | 0.08 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr18_+_13218769 | 0.08 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_125496619 | 0.08 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr12_+_25205628 | 0.08 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr11_-_82708435 | 0.08 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr3_+_181429704 | 0.08 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr18_+_3451646 | 0.08 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr16_+_30406423 | 0.08 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr5_+_140254884 | 0.08 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr3_+_52719936 | 0.08 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr19_-_59030921 | 0.08 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr12_+_9066472 | 0.08 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr11_-_125365435 | 0.08 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr11_-_76155618 | 0.08 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr4_-_109089573 | 0.08 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr12_-_8815215 | 0.08 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr11_+_46402583 | 0.08 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr12_-_104359475 | 0.07 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr4_-_109090106 | 0.07 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr4_+_54243862 | 0.07 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr22_+_32439019 | 0.07 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_-_8892464 | 0.07 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr21_-_40720995 | 0.07 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr11_-_79151695 | 0.07 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr10_-_13390270 | 0.07 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr6_+_32937083 | 0.07 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr2_-_208890218 | 0.07 |
ENST00000457206.1
ENST00000427836.2 ENST00000389247.4 |
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3 |
| chr5_+_140792614 | 0.07 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr2_-_204399976 | 0.07 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr17_+_9066252 | 0.07 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr1_+_14075865 | 0.06 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr4_+_89300158 | 0.06 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_+_140235469 | 0.06 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr11_-_82708519 | 0.06 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr11_+_63754294 | 0.06 |
ENST00000543988.1
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr15_+_80696666 | 0.06 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr16_+_23765948 | 0.06 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr11_+_63753883 | 0.06 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr17_+_30813576 | 0.06 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr17_-_76719638 | 0.06 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr7_-_150924121 | 0.06 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr15_+_57211318 | 0.06 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr2_-_70475586 | 0.06 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_+_67498396 | 0.06 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_+_140593509 | 0.06 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr8_+_42195972 | 0.06 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr1_-_151431674 | 0.06 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr5_+_140213815 | 0.06 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr14_-_91884150 | 0.06 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr4_-_186877481 | 0.06 |
ENST00000444781.1
ENST00000432655.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_64013663 | 0.06 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_62414070 | 0.06 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr16_+_89989687 | 0.06 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr9_+_112852477 | 0.06 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr1_+_47799542 | 0.06 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr2_+_65215604 | 0.06 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chrX_+_135579670 | 0.05 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr14_+_103851712 | 0.05 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr3_+_39509163 | 0.05 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr19_+_49468558 | 0.05 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr2_-_47572207 | 0.05 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr18_+_55102917 | 0.05 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr3_-_8686479 | 0.05 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr7_+_139528952 | 0.05 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr12_-_109219937 | 0.05 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr11_+_131781290 | 0.05 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr2_-_207078086 | 0.05 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr18_-_52989217 | 0.05 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr7_+_87563557 | 0.05 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr12_+_32260085 | 0.05 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr6_-_24877490 | 0.05 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr7_-_107643567 | 0.05 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr2_-_70475730 | 0.05 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_+_21729593 | 0.05 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr5_-_176037105 | 0.05 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr20_-_3996036 | 0.05 |
ENST00000336095.6
|
RNF24
|
ring finger protein 24 |
| chr12_-_18890940 | 0.05 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr17_+_1945301 | 0.05 |
ENST00000572195.1
|
OVCA2
|
ovarian tumor suppressor candidate 2 |
| chr3_-_57583130 | 0.05 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr8_+_11660227 | 0.05 |
ENST00000443614.2
ENST00000525900.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr15_-_54025300 | 0.05 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr6_-_127840021 | 0.05 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr4_-_185726906 | 0.05 |
ENST00000513317.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_+_55524085 | 0.05 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr3_+_39509070 | 0.05 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr20_-_20693131 | 0.05 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr12_-_58145604 | 0.05 |
ENST00000552254.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr16_+_54964740 | 0.05 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr19_-_18717627 | 0.05 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr1_-_26232951 | 0.05 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr5_-_157002775 | 0.05 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chrX_-_117107542 | 0.04 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr12_-_58146128 | 0.04 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr11_+_46402744 | 0.04 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_-_109776901 | 0.04 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_85641162 | 0.04 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chrX_+_70503433 | 0.04 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr19_+_58694742 | 0.04 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr1_-_212004090 | 0.04 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_+_61293021 | 0.04 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr21_+_33784670 | 0.04 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr10_+_123923105 | 0.04 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chrX_+_12993202 | 0.04 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr1_+_249200395 | 0.04 |
ENST00000355360.4
ENST00000329291.5 ENST00000539153.1 |
PGBD2
|
piggyBac transposable element derived 2 |
| chr15_+_79166065 | 0.04 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr10_+_35415851 | 0.04 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr10_-_128975273 | 0.04 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr21_-_33104367 | 0.04 |
ENST00000286835.7
ENST00000399804.1 |
SCAF4
|
SR-related CTD-associated factor 4 |
| chr9_-_98269481 | 0.04 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr7_+_73868439 | 0.04 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr7_-_107643674 | 0.04 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr1_-_207143802 | 0.04 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr11_-_44971702 | 0.04 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr12_-_45269769 | 0.04 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr2_-_225266743 | 0.04 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr1_-_32801825 | 0.04 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr7_+_99699179 | 0.04 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr10_+_104535994 | 0.04 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr14_+_100070869 | 0.04 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr12_-_109797249 | 0.04 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr15_+_90744745 | 0.04 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr6_-_29527702 | 0.04 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr18_+_3449821 | 0.04 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_134639180 | 0.04 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chrX_+_65384052 | 0.04 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr16_+_30406721 | 0.04 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr12_+_50479101 | 0.04 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_-_6479963 | 0.04 |
ENST00000377836.4
ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2
|
hes family bHLH transcription factor 2 |
| chr4_-_140477910 | 0.04 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chrX_-_106959631 | 0.04 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr6_+_13925098 | 0.04 |
ENST00000488300.1
ENST00000544682.1 ENST00000420478.2 |
RNF182
|
ring finger protein 182 |
| chr1_-_26324534 | 0.04 |
ENST00000374284.1
ENST00000441420.1 ENST00000374282.3 |
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr12_-_105478339 | 0.04 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr7_+_99006232 | 0.03 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr8_+_11660120 | 0.03 |
ENST00000220584.4
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr22_+_38004723 | 0.03 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr12_+_50898881 | 0.03 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr10_+_24528108 | 0.03 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr11_+_64949343 | 0.03 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr3_+_112930306 | 0.03 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.0 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |