Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
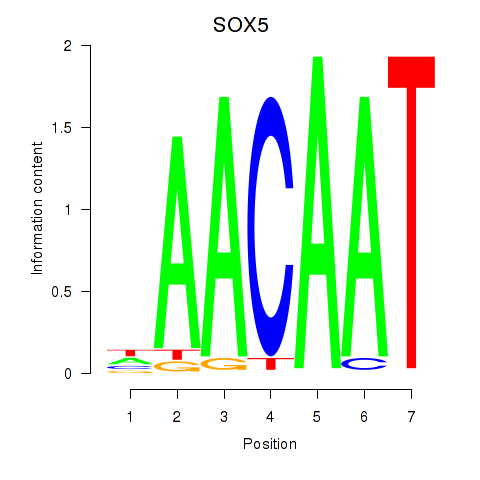
Results for SOX5
Z-value: 0.93
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg19_v2_chr12_-_24103954_24103972 | -0.04 | 9.4e-01 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_166702601 | 0.43 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr17_+_33448593 | 0.38 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr14_-_92413727 | 0.36 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr2_-_164592497 | 0.34 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr3_+_112930306 | 0.31 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr17_+_67498396 | 0.28 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr3_+_25469802 | 0.27 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr3_+_181429704 | 0.27 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr19_+_36132631 | 0.26 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr5_-_146461027 | 0.25 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_25451065 | 0.24 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr4_+_165675197 | 0.24 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr8_-_72274467 | 0.24 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr3_+_25469724 | 0.24 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr7_+_116451100 | 0.23 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr9_-_98268883 | 0.23 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr7_-_83824449 | 0.23 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_219615984 | 0.21 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr6_-_134861089 | 0.21 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr1_-_167883327 | 0.21 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr1_-_40367668 | 0.20 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr17_+_30814707 | 0.20 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr19_-_7698599 | 0.20 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr3_+_89156799 | 0.19 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr6_+_122720681 | 0.19 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr9_-_115249484 | 0.19 |
ENST00000457681.1
|
C9orf147
|
chromosome 9 open reading frame 147 |
| chr1_-_204463829 | 0.19 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr9_-_72287191 | 0.18 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr2_-_70475586 | 0.18 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr3_-_123339418 | 0.18 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr21_+_35553045 | 0.18 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chrX_-_106959631 | 0.18 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr15_+_49715449 | 0.18 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr14_-_92414294 | 0.18 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr20_-_52612468 | 0.17 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr4_+_95916947 | 0.17 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr4_+_11627248 | 0.17 |
ENST00000510095.1
|
RP11-281P23.2
|
RP11-281P23.2 |
| chr11_+_46402482 | 0.17 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_-_39459103 | 0.16 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr1_-_243349684 | 0.16 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_+_205682497 | 0.16 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr12_-_109797249 | 0.16 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr8_-_72274355 | 0.15 |
ENST00000388741.2
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrX_-_50557302 | 0.15 |
ENST00000289292.7
|
SHROOM4
|
shroom family member 4 |
| chr2_-_192711968 | 0.14 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr2_-_188312971 | 0.14 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr12_-_96793142 | 0.14 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr15_-_34610962 | 0.14 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr7_-_11871815 | 0.13 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr1_-_40367530 | 0.13 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr10_+_99079008 | 0.13 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr4_+_165675269 | 0.13 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chrX_-_117107542 | 0.13 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr12_+_498500 | 0.13 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr20_-_3762087 | 0.13 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr1_+_33722080 | 0.13 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr17_+_1666108 | 0.13 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_+_91416092 | 0.13 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr15_+_84115868 | 0.13 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_+_110958124 | 0.12 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr15_+_49715293 | 0.12 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr5_+_67588391 | 0.12 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_53443963 | 0.12 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr6_+_155537771 | 0.12 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr15_+_90319557 | 0.12 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr3_-_123339343 | 0.12 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr12_+_53443680 | 0.12 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr14_-_73360796 | 0.12 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr1_-_12677714 | 0.12 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chrX_+_9880412 | 0.12 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr10_-_116444371 | 0.12 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr4_+_110749143 | 0.11 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr2_+_47454054 | 0.11 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr19_+_18794470 | 0.11 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chrX_-_50557014 | 0.11 |
ENST00000376020.2
|
SHROOM4
|
shroom family member 4 |
| chr6_-_79944336 | 0.11 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr20_-_61002584 | 0.11 |
ENST00000252998.1
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr10_-_14614311 | 0.11 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_-_53018654 | 0.11 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr12_-_111926342 | 0.11 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr1_+_164528866 | 0.11 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_25657444 | 0.11 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr1_-_50889155 | 0.10 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr11_-_82708435 | 0.10 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr10_-_14614122 | 0.10 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_-_92414055 | 0.10 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr6_-_32557610 | 0.10 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr7_+_111846741 | 0.10 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr14_+_61654271 | 0.10 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr2_+_30569506 | 0.10 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr10_-_14613968 | 0.10 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr9_+_137218362 | 0.10 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr9_-_15510989 | 0.10 |
ENST00000380715.1
ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chrX_+_24483338 | 0.10 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr12_-_12674032 | 0.10 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr9_-_134145880 | 0.09 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr13_+_22245522 | 0.09 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr3_+_89156674 | 0.09 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr17_-_47786375 | 0.09 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr6_-_56716686 | 0.09 |
ENST00000520645.1
|
DST
|
dystonin |
| chr11_-_72852958 | 0.09 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr4_-_184241927 | 0.09 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr12_-_122985067 | 0.09 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr1_+_164529004 | 0.09 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr18_+_3451646 | 0.09 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_+_50479109 | 0.09 |
ENST00000550477.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr18_+_21594585 | 0.09 |
ENST00000317571.3
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr11_+_46402583 | 0.09 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr9_-_14314518 | 0.08 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr8_-_72274095 | 0.08 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr9_-_126030817 | 0.08 |
ENST00000348403.5
ENST00000447404.2 ENST00000360998.3 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr20_-_52612705 | 0.08 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_-_156217829 | 0.08 |
ENST00000356983.2
ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr3_+_171561127 | 0.08 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr10_-_99094458 | 0.08 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr4_+_39408470 | 0.08 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr5_+_140235469 | 0.08 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr18_+_3451584 | 0.08 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_-_123476612 | 0.08 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr12_+_9066472 | 0.08 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chrX_-_117107680 | 0.08 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr9_-_14314566 | 0.08 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr6_+_126070726 | 0.08 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr8_-_57123815 | 0.08 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr10_-_14614095 | 0.08 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr7_-_32931623 | 0.07 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chrX_-_112084043 | 0.07 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr19_+_31658405 | 0.07 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr3_-_11685345 | 0.07 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr12_-_121973974 | 0.07 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr11_-_79151695 | 0.07 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr14_+_37126765 | 0.07 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr12_+_50479101 | 0.07 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr9_-_123476719 | 0.07 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr5_+_138678131 | 0.07 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr4_-_186732048 | 0.07 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr21_-_39870339 | 0.07 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr14_-_69619823 | 0.07 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr11_+_46402744 | 0.07 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_-_13290684 | 0.07 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr1_+_147013182 | 0.07 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr1_-_156217822 | 0.07 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr10_-_62332357 | 0.07 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr18_-_53177984 | 0.06 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr2_-_230787879 | 0.06 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr4_+_170581213 | 0.06 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr16_+_86612112 | 0.06 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr9_-_92051354 | 0.06 |
ENST00000418828.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr4_-_186732892 | 0.06 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_129097479 | 0.06 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr1_-_243418650 | 0.06 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr11_+_19799327 | 0.06 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr18_-_52989217 | 0.06 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_-_102323740 | 0.06 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr1_-_176176114 | 0.06 |
ENST00000308769.8
|
RFWD2
|
ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
| chr18_+_20494078 | 0.06 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr12_+_32654965 | 0.06 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr15_+_57211318 | 0.05 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr10_+_63661053 | 0.05 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr14_-_22005018 | 0.05 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_156217875 | 0.05 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr10_+_121578211 | 0.05 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr15_-_52944231 | 0.05 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr1_-_11751529 | 0.05 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr12_-_122985494 | 0.05 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr6_+_43027595 | 0.05 |
ENST00000259708.3
ENST00000472792.1 ENST00000479388.1 ENST00000460283.1 ENST00000394056.2 |
KLC4
|
kinesin light chain 4 |
| chr12_-_92536433 | 0.05 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr22_-_36236265 | 0.05 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_18466026 | 0.05 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr4_-_87281196 | 0.05 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_+_61547405 | 0.05 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr9_+_71394945 | 0.05 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr3_-_141747439 | 0.05 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_-_62149433 | 0.05 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_29241027 | 0.05 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr2_-_70475701 | 0.05 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr21_-_40720995 | 0.05 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr13_+_103459704 | 0.05 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr5_+_173316341 | 0.05 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr9_+_118916082 | 0.05 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr3_-_19988462 | 0.05 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr11_+_19798964 | 0.05 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chrX_+_15525426 | 0.05 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr18_-_52989525 | 0.05 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_+_120315160 | 0.05 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr6_+_37137939 | 0.05 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr1_-_11751665 | 0.04 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr17_+_67498295 | 0.04 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr3_+_154797428 | 0.04 |
ENST00000460393.1
|
MME
|
membrane metallo-endopeptidase |
| chr12_+_498545 | 0.04 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr14_-_69619689 | 0.04 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr14_-_69619291 | 0.04 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr12_-_99288536 | 0.04 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_-_69215467 | 0.04 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1 |
| chr18_+_21594384 | 0.04 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr16_+_20911174 | 0.04 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr2_+_203499901 | 0.04 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr11_-_82708519 | 0.04 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr9_-_124989804 | 0.04 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr12_+_56360550 | 0.04 |
ENST00000266970.4
|
CDK2
|
cyclin-dependent kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.3 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.0 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |