Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
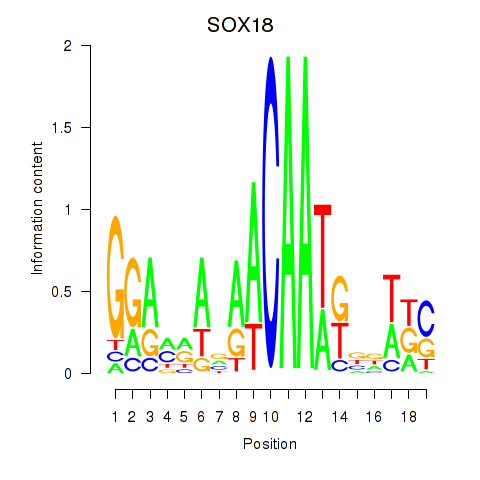
Results for SOX18
Z-value: 0.73
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | -0.27 | 6.1e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_42192189 | 0.61 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr1_+_37947257 | 0.42 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr2_+_101591314 | 0.41 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_+_86999516 | 0.41 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr5_-_146461027 | 0.40 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_+_41158742 | 0.40 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr10_-_4285923 | 0.36 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr8_+_40010989 | 0.34 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr6_-_133035185 | 0.33 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr19_-_12708113 | 0.32 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr5_+_127039075 | 0.30 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr17_-_76719638 | 0.29 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr2_-_114647327 | 0.29 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr19_-_42006539 | 0.28 |
ENST00000597702.1
ENST00000588495.1 ENST00000595837.1 ENST00000594315.1 |
AC011526.1
|
AC011526.1 |
| chr12_+_113344582 | 0.26 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_117021430 | 0.25 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr15_-_96590126 | 0.25 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr22_-_38480100 | 0.25 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr12_-_10573149 | 0.24 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr19_+_36132631 | 0.24 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr1_+_206972215 | 0.23 |
ENST00000340758.2
|
IL19
|
interleukin 19 |
| chr6_-_26235206 | 0.22 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr12_-_11139511 | 0.20 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr11_+_5710919 | 0.20 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr7_-_98805129 | 0.19 |
ENST00000327442.6
|
KPNA7
|
karyopherin alpha 7 (importin alpha 8) |
| chr11_-_76155618 | 0.19 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr6_+_24126350 | 0.18 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr7_-_117512264 | 0.18 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chrM_+_8366 | 0.18 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr17_-_39123144 | 0.18 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr8_+_119294456 | 0.17 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr4_-_76944621 | 0.17 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr22_+_20850171 | 0.17 |
ENST00000445987.1
|
MED15
|
mediator complex subunit 15 |
| chr12_+_104697504 | 0.17 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr11_+_5711010 | 0.17 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr1_+_55271736 | 0.17 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr6_+_26217159 | 0.16 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr3_-_146262637 | 0.16 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr4_+_156588115 | 0.15 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr1_+_153003671 | 0.15 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr22_-_31324215 | 0.14 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr7_+_141408153 | 0.14 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr11_+_71238313 | 0.14 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr19_-_39805976 | 0.13 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr13_-_44735393 | 0.13 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr16_-_79804394 | 0.13 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr17_+_38516907 | 0.13 |
ENST00000578774.1
|
CTD-2267D19.3
|
Uncharacterized protein |
| chr19_+_107471 | 0.13 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr4_-_100484825 | 0.13 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr12_+_70574088 | 0.13 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr22_+_45714672 | 0.12 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr2_-_208890218 | 0.12 |
ENST00000457206.1
ENST00000427836.2 ENST00000389247.4 |
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3 |
| chr6_-_88032795 | 0.12 |
ENST00000296882.3
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr22_+_31488433 | 0.12 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr20_+_31870927 | 0.11 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chrX_+_47078069 | 0.11 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr1_+_172745006 | 0.11 |
ENST00000432694.2
|
RP1-15D23.2
|
RP1-15D23.2 |
| chr18_+_3447572 | 0.11 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_30899924 | 0.11 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr5_+_32711419 | 0.11 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_-_32801825 | 0.11 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr19_+_41256764 | 0.11 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_105953972 | 0.10 |
ENST00000410049.1
|
C2orf49
|
chromosome 2 open reading frame 49 |
| chr6_-_134639180 | 0.10 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_41166414 | 0.10 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chrX_+_47077632 | 0.10 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr10_-_105992059 | 0.10 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr11_-_89224299 | 0.10 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr2_-_153573965 | 0.09 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr12_+_110940111 | 0.09 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr4_+_54243798 | 0.09 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr7_-_45026419 | 0.09 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr5_-_31532238 | 0.09 |
ENST00000507438.1
|
DROSHA
|
drosha, ribonuclease type III |
| chr12_+_96252706 | 0.09 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr3_-_122283424 | 0.09 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_+_113258495 | 0.09 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr15_+_75628394 | 0.09 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr4_+_69962212 | 0.09 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_+_225600404 | 0.09 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr3_+_186330712 | 0.09 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr15_+_75628232 | 0.09 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr3_+_188817891 | 0.09 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr12_+_32260085 | 0.09 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr6_-_88001706 | 0.08 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr11_-_75236867 | 0.08 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr11_-_111749767 | 0.08 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_+_66258846 | 0.08 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_11002063 | 0.08 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chrM_+_8527 | 0.08 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr6_-_128841503 | 0.08 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr7_+_157129660 | 0.08 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr17_-_8661860 | 0.08 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr6_-_31125850 | 0.08 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr8_-_123793048 | 0.08 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr11_-_69867159 | 0.08 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr15_+_44084040 | 0.08 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr8_-_37707356 | 0.08 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr18_-_5396271 | 0.08 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr1_+_10459433 | 0.08 |
ENST00000465632.1
ENST00000460189.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr7_-_45026200 | 0.08 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr6_+_155334780 | 0.08 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr12_-_10562356 | 0.08 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr1_-_182361327 | 0.08 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr1_+_209929377 | 0.08 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr8_+_35649365 | 0.07 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr12_+_69633372 | 0.07 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_-_11150474 | 0.07 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr3_+_108541545 | 0.07 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr13_-_46716969 | 0.07 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr13_-_46756351 | 0.07 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_-_99716940 | 0.07 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_54893250 | 0.07 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr19_-_42192096 | 0.07 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr17_+_68071389 | 0.07 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_-_89224508 | 0.07 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chrX_+_85969626 | 0.07 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr9_+_137987825 | 0.07 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr6_-_110500905 | 0.07 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr5_+_57878859 | 0.07 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr19_-_43382142 | 0.07 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr8_+_24151620 | 0.07 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr5_-_43515125 | 0.07 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr12_-_85430024 | 0.07 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr1_+_43124087 | 0.07 |
ENST00000304979.3
ENST00000372550.1 ENST00000440068.1 |
PPIH
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr17_+_6658878 | 0.07 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr15_+_67418047 | 0.07 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr19_+_50183032 | 0.07 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr13_-_33780133 | 0.07 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr4_-_144940477 | 0.07 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chrX_+_52920336 | 0.06 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr2_+_210518057 | 0.06 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr19_+_4402659 | 0.06 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_1418154 | 0.06 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr11_-_58275578 | 0.06 |
ENST00000360374.2
|
OR5B21
|
olfactory receptor, family 5, subfamily B, member 21 |
| chr12_+_69633317 | 0.06 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr13_-_24471641 | 0.06 |
ENST00000382145.1
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr3_+_186435065 | 0.06 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr19_+_39971505 | 0.06 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr17_+_12569472 | 0.06 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr7_-_6048650 | 0.06 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr10_-_13390270 | 0.06 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr7_-_148725733 | 0.06 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr2_+_241544834 | 0.06 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr11_+_134201911 | 0.06 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chrX_+_10031499 | 0.06 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_89224488 | 0.05 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr11_-_89224139 | 0.05 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr8_+_7397150 | 0.05 |
ENST00000533250.1
|
RP11-1118M6.1
|
proline rich 23 domain containing 1 |
| chr11_+_73358690 | 0.05 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_-_73497581 | 0.05 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr11_+_74699942 | 0.05 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr15_+_75628419 | 0.05 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr4_+_156588249 | 0.05 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr11_-_111749878 | 0.05 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_+_161123536 | 0.05 |
ENST00000368003.5
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr20_+_31499459 | 0.05 |
ENST00000439060.1
|
EFCAB8
|
EF-hand calcium binding domain 8 |
| chr2_-_165811756 | 0.05 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr2_-_100939195 | 0.05 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr3_-_197300194 | 0.05 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_+_68071458 | 0.05 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_+_28624878 | 0.05 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr11_+_64949343 | 0.05 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr1_-_206785898 | 0.05 |
ENST00000271764.2
|
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr3_-_121379739 | 0.05 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr19_+_44576296 | 0.05 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284 |
| chr12_-_8815215 | 0.05 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr5_-_55412774 | 0.05 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr8_-_7638935 | 0.05 |
ENST00000528972.1
|
AC084121.16
|
proline rich 23 domain containing 2 |
| chr9_+_125437315 | 0.05 |
ENST00000304820.2
|
OR1L3
|
olfactory receptor, family 1, subfamily L, member 3 |
| chr17_+_6916527 | 0.05 |
ENST00000552321.1
|
RNASEK
|
ribonuclease, RNase K |
| chr9_-_127533519 | 0.04 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr2_-_47382414 | 0.04 |
ENST00000294947.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr11_+_64949899 | 0.04 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr12_-_11422739 | 0.04 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chrX_-_110507098 | 0.04 |
ENST00000541758.1
|
CAPN6
|
calpain 6 |
| chr19_+_8117881 | 0.04 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr18_+_32455201 | 0.04 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr2_+_217277271 | 0.04 |
ENST00000425815.1
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr4_+_88529681 | 0.04 |
ENST00000399271.1
|
DSPP
|
dentin sialophosphoprotein |
| chr11_+_20620946 | 0.04 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr17_+_8316442 | 0.04 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr11_-_89223883 | 0.04 |
ENST00000528341.1
|
NOX4
|
NADPH oxidase 4 |
| chr3_+_37034823 | 0.04 |
ENST00000231790.2
ENST00000456676.2 |
MLH1
|
mutL homolog 1 |
| chr7_+_141811539 | 0.04 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr5_+_142125161 | 0.04 |
ENST00000432677.1
|
AC005592.1
|
AC005592.1 |
| chr11_-_104817919 | 0.04 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr21_-_15918618 | 0.04 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr12_-_56321397 | 0.04 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr12_-_75603202 | 0.04 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chrX_+_2984874 | 0.04 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr2_-_40739501 | 0.03 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_+_8021713 | 0.03 |
ENST00000338639.5
ENST00000493678.1 ENST00000377493.5 |
PARK7
|
parkinson protein 7 |
| chr3_-_120003941 | 0.03 |
ENST00000464295.1
|
GPR156
|
G protein-coupled receptor 156 |
| chr19_+_41257084 | 0.03 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_-_32908792 | 0.03 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr12_-_49999389 | 0.03 |
ENST00000551047.1
ENST00000544141.1 |
FAM186B
|
family with sequence similarity 186, member B |
| chr3_-_112693865 | 0.03 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr1_-_206785789 | 0.03 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr12_-_70093190 | 0.03 |
ENST00000330891.5
|
BEST3
|
bestrophin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.2 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.3 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.2 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.3 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.0 | GO:0050787 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.0 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |