Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SOX17
Z-value: 0.27
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SRY-box transcription factor 17 |
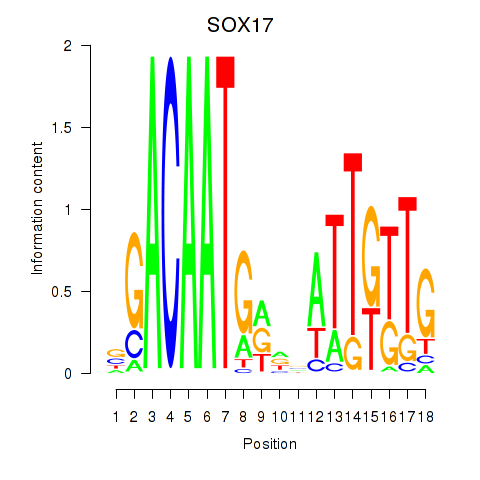
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_94577074 | 0.65 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr4_-_74864386 | 0.50 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr11_+_18154059 | 0.34 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr17_+_41158742 | 0.19 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr6_+_31916733 | 0.18 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr11_+_107650219 | 0.15 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr19_+_41882466 | 0.13 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr9_+_99690592 | 0.13 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr20_-_62199427 | 0.13 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr11_-_125365435 | 0.11 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_+_158901329 | 0.11 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr6_+_126240442 | 0.10 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_-_125366018 | 0.10 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr19_+_45690646 | 0.09 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr19_+_16607122 | 0.09 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr12_+_498545 | 0.09 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr19_-_13710678 | 0.08 |
ENST00000592864.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr19_-_35454953 | 0.08 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr14_-_105531759 | 0.08 |
ENST00000329797.3
ENST00000539291.2 ENST00000392585.2 |
GPR132
|
G protein-coupled receptor 132 |
| chr8_+_104383728 | 0.08 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr16_-_90142338 | 0.08 |
ENST00000407825.1
ENST00000449207.2 |
PRDM7
|
PR domain containing 7 |
| chr11_-_125351481 | 0.08 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr18_+_22040593 | 0.08 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr19_+_56653064 | 0.07 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr7_+_112120908 | 0.07 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr14_+_77882741 | 0.07 |
ENST00000595520.1
|
FKSG61
|
FKSG61 |
| chr14_+_39703112 | 0.07 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr5_+_171621176 | 0.07 |
ENST00000398186.4
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr17_-_41132410 | 0.07 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr19_-_44809121 | 0.06 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr22_+_24951949 | 0.06 |
ENST00000402849.1
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr15_-_81616446 | 0.06 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_-_115818951 | 0.06 |
ENST00000553380.1
ENST00000374227.3 |
ZFP37
|
ZFP37 zinc finger protein |
| chr9_-_115819039 | 0.06 |
ENST00000555206.1
|
ZFP37
|
ZFP37 zinc finger protein |
| chr11_-_67141090 | 0.06 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr5_+_142286887 | 0.06 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr16_-_71523236 | 0.06 |
ENST00000288177.5
ENST00000569072.1 |
ZNF19
|
zinc finger protein 19 |
| chr17_-_36997708 | 0.06 |
ENST00000398575.4
|
C17orf98
|
chromosome 17 open reading frame 98 |
| chr15_-_65117807 | 0.06 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr8_-_19615382 | 0.06 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr3_+_142842128 | 0.06 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr9_-_97090926 | 0.06 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr1_-_206306107 | 0.06 |
ENST00000436158.1
ENST00000455672.1 |
RP11-38J22.6
|
RP11-38J22.6 |
| chr11_-_102668879 | 0.06 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr20_-_45318230 | 0.06 |
ENST00000372114.3
|
TP53RK
|
TP53 regulating kinase |
| chr6_-_133079022 | 0.06 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr8_-_66474884 | 0.06 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr4_-_122085469 | 0.06 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr6_-_112081113 | 0.06 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr5_+_127039075 | 0.06 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr8_-_95220775 | 0.06 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr2_-_118943930 | 0.06 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr7_+_137761220 | 0.05 |
ENST00000242375.3
ENST00000438242.1 |
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr13_-_30951282 | 0.05 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr18_-_70931689 | 0.05 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr18_-_43684186 | 0.05 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr11_-_7695437 | 0.05 |
ENST00000533558.1
ENST00000527542.1 ENST00000531096.1 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr7_+_137761167 | 0.05 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr14_+_22591276 | 0.05 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr6_-_42858534 | 0.05 |
ENST00000408925.2
|
C6orf226
|
chromosome 6 open reading frame 226 |
| chr7_+_100551239 | 0.05 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr11_+_60145948 | 0.05 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr2_-_86094764 | 0.05 |
ENST00000393808.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr10_-_90611566 | 0.05 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr6_-_26216872 | 0.05 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr20_-_43883197 | 0.05 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr5_-_146833222 | 0.05 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chrX_-_118699325 | 0.05 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr10_-_112255945 | 0.05 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr15_-_43212996 | 0.05 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr14_-_55369525 | 0.05 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr20_-_1309809 | 0.05 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr1_+_10509971 | 0.05 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr14_-_75083313 | 0.04 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr1_-_201476274 | 0.04 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr15_+_77287426 | 0.04 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr1_+_95975672 | 0.04 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr17_-_57784755 | 0.04 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr11_+_73000449 | 0.04 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr22_+_24951436 | 0.04 |
ENST00000215829.3
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa |
| chr19_+_10765003 | 0.04 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr19_-_48753028 | 0.04 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_+_29181503 | 0.04 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr16_-_71523179 | 0.04 |
ENST00000564230.1
ENST00000565637.1 |
ZNF19
|
zinc finger protein 19 |
| chr4_+_100495864 | 0.04 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr19_+_10764937 | 0.04 |
ENST00000449870.1
ENST00000318511.3 ENST00000420083.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr19_+_34895289 | 0.04 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr1_+_227751281 | 0.04 |
ENST00000440339.1
|
ZNF678
|
zinc finger protein 678 |
| chr10_+_135340859 | 0.04 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr2_+_113816215 | 0.04 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr19_+_40877583 | 0.04 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr7_+_16828866 | 0.04 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr2_+_48844937 | 0.04 |
ENST00000448460.1
ENST00000437125.1 ENST00000430487.2 |
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr18_+_9885760 | 0.04 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr12_-_15374328 | 0.04 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr7_-_150924121 | 0.04 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr6_-_49755019 | 0.04 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr16_+_3704822 | 0.04 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr20_-_44207965 | 0.04 |
ENST00000357199.4
ENST00000289953.2 |
WFDC8
|
WAP four-disulfide core domain 8 |
| chr21_-_16125773 | 0.03 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr20_+_36405665 | 0.03 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr5_-_138842286 | 0.03 |
ENST00000515823.1
|
ECSCR
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr20_-_45318051 | 0.03 |
ENST00000372102.3
|
TP53RK
|
TP53 regulating kinase |
| chr2_-_100939195 | 0.03 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr19_-_10764509 | 0.03 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr15_-_43212836 | 0.03 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chrX_+_70503037 | 0.03 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr19_-_55720824 | 0.03 |
ENST00000376350.3
ENST00000263434.5 |
PTPRH
|
protein tyrosine phosphatase, receptor type, H |
| chr7_-_130597935 | 0.03 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr15_-_28778117 | 0.03 |
ENST00000525590.2
ENST00000329523.6 |
GOLGA8G
|
golgin A8 family, member G |
| chr10_-_91295304 | 0.03 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr3_-_15540055 | 0.03 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr2_-_114205429 | 0.03 |
ENST00000440617.1
|
AC016745.1
|
HDCMB45P; Uncharacterized protein |
| chr18_+_1099004 | 0.03 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1 |
| chr2_+_48844973 | 0.03 |
ENST00000403751.3
|
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr9_-_70866572 | 0.03 |
ENST00000417734.1
|
AL353608.1
|
HDCMB45P; Uncharacterized protein |
| chr10_-_10836865 | 0.03 |
ENST00000446372.2
|
SFTA1P
|
surfactant associated 1, pseudogene |
| chr8_+_133931648 | 0.03 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr17_-_4852243 | 0.03 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr4_+_155665123 | 0.03 |
ENST00000336356.3
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr19_-_7167989 | 0.03 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr4_-_69083720 | 0.03 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr5_+_49962772 | 0.03 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_94300474 | 0.03 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr18_-_43678241 | 0.03 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr2_+_157330081 | 0.03 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr3_-_151176497 | 0.03 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr19_+_107471 | 0.03 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr10_-_135379132 | 0.03 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr10_-_128975273 | 0.03 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr7_+_137761199 | 0.03 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr3_+_174577070 | 0.03 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr16_-_8622226 | 0.03 |
ENST00000568335.1
|
TMEM114
|
transmembrane protein 114 |
| chr3_-_52488048 | 0.03 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr8_-_52721975 | 0.03 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr4_-_100065419 | 0.03 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr10_+_6622345 | 0.03 |
ENST00000445427.1
ENST00000455810.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr1_+_149239529 | 0.03 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chrX_+_129473916 | 0.03 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr18_+_3451584 | 0.03 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_24969755 | 0.03 |
ENST00000447431.2
ENST00000374389.4 |
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr18_+_5238055 | 0.03 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr8_-_19615435 | 0.03 |
ENST00000523262.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr5_-_1345199 | 0.03 |
ENST00000320895.5
|
CLPTM1L
|
CLPTM1-like |
| chr18_-_24283586 | 0.03 |
ENST00000579458.1
|
U3
|
Small nucleolar RNA U3 |
| chrX_+_47083037 | 0.03 |
ENST00000523034.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr11_-_74022658 | 0.03 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr11_-_59950622 | 0.03 |
ENST00000323961.3
ENST00000412309.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr10_-_52000307 | 0.02 |
ENST00000443575.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr6_+_53964336 | 0.02 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chr4_-_170897045 | 0.02 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr18_+_21269404 | 0.02 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_10163231 | 0.02 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr17_+_58042422 | 0.02 |
ENST00000586209.1
|
RP11-178C3.2
|
RP11-178C3.2 |
| chr2_+_232573222 | 0.02 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr3_+_88108381 | 0.02 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr19_-_48752812 | 0.02 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr19_+_58095501 | 0.02 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr4_-_141348763 | 0.02 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr15_+_88120158 | 0.02 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr2_+_239335449 | 0.02 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr5_+_112196919 | 0.02 |
ENST00000505459.1
ENST00000282999.3 ENST00000515463.1 |
SRP19
|
signal recognition particle 19kDa |
| chr1_+_40840320 | 0.02 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr12_+_9822331 | 0.02 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr22_-_32058166 | 0.02 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr2_+_10262857 | 0.02 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr4_-_100065389 | 0.02 |
ENST00000512499.1
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr7_+_139026057 | 0.02 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr8_-_19615538 | 0.02 |
ENST00000517494.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr1_+_197881592 | 0.02 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr2_+_103035102 | 0.02 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr3_-_128207349 | 0.02 |
ENST00000487848.1
|
GATA2
|
GATA binding protein 2 |
| chrY_-_20935572 | 0.02 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr3_+_185303962 | 0.02 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr16_+_2867228 | 0.02 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr2_+_201677390 | 0.02 |
ENST00000447069.1
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr2_-_157186630 | 0.02 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr13_+_25338290 | 0.02 |
ENST00000255324.5
ENST00000381921.1 ENST00000255325.6 |
RNF17
|
ring finger protein 17 |
| chr4_-_15939963 | 0.02 |
ENST00000259988.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr1_+_46016703 | 0.02 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr1_+_207669495 | 0.02 |
ENST00000367052.1
ENST00000367051.1 ENST00000367053.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr1_-_153917700 | 0.02 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr19_-_48753064 | 0.02 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr19_-_47735918 | 0.02 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr2_+_235903480 | 0.02 |
ENST00000454947.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr12_-_96794143 | 0.02 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr17_+_41132564 | 0.02 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chrY_+_20708557 | 0.02 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr2_+_102413726 | 0.02 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_-_45838011 | 0.02 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr14_-_36645674 | 0.02 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr8_+_72587535 | 0.02 |
ENST00000519840.1
ENST00000521131.1 |
RP11-1144P22.1
|
RP11-1144P22.1 |
| chr11_-_57417405 | 0.02 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr13_+_24144796 | 0.02 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr8_+_27631903 | 0.02 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr2_+_33661382 | 0.02 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr16_+_22517166 | 0.02 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr7_-_107642348 | 0.02 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr9_-_100935043 | 0.02 |
ENST00000343933.5
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr1_+_81106951 | 0.02 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.0 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |