Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
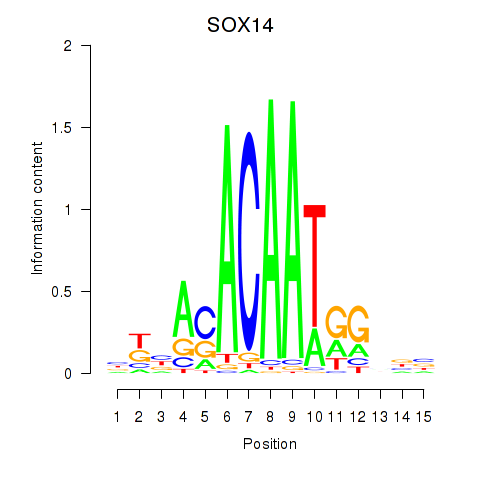
Results for SOX14
Z-value: 0.26
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Activity profile of SOX14 motif
Sorted Z-values of SOX14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_111755280 | 0.25 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr19_-_48673465 | 0.25 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr12_+_65996599 | 0.20 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr3_-_171489085 | 0.13 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr17_-_39677971 | 0.11 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr6_+_122720681 | 0.11 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr2_-_55276320 | 0.11 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chrX_-_106959631 | 0.10 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr17_+_67498396 | 0.10 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_-_15872914 | 0.10 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_240323439 | 0.09 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr5_+_95066823 | 0.09 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr3_+_48507210 | 0.08 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr15_+_71184931 | 0.08 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr19_-_5340730 | 0.07 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr1_+_52521928 | 0.07 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr6_-_86353510 | 0.07 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr12_+_98909520 | 0.07 |
ENST00000261210.5
|
TMPO
|
thymopoietin |
| chr4_-_120548779 | 0.07 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr2_+_54350316 | 0.07 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr1_+_65886326 | 0.07 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr3_-_101232019 | 0.07 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr17_+_7608511 | 0.07 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr2_-_166702601 | 0.07 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr11_+_17316870 | 0.07 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr1_-_159915386 | 0.07 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr6_+_127898312 | 0.07 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr3_+_51851612 | 0.07 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr1_-_204463829 | 0.06 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr12_+_498500 | 0.06 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr17_-_18585131 | 0.06 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chrX_-_138914394 | 0.06 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr14_+_76776957 | 0.06 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr3_-_18466026 | 0.06 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr2_-_106015527 | 0.06 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr1_+_52521797 | 0.06 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr1_+_201857798 | 0.06 |
ENST00000362011.6
|
SHISA4
|
shisa family member 4 |
| chrX_+_129473859 | 0.06 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr12_+_69004619 | 0.06 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr17_+_64961026 | 0.06 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr14_+_52313833 | 0.06 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_134551583 | 0.05 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr5_-_180018540 | 0.05 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chrX_+_12993336 | 0.05 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr7_+_121513374 | 0.05 |
ENST00000449182.1
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr4_+_113152978 | 0.05 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_170590321 | 0.05 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr3_+_186649133 | 0.05 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr2_+_127413677 | 0.05 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr12_-_15942503 | 0.05 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_1266654 | 0.05 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr1_+_226250379 | 0.05 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr1_+_209602156 | 0.05 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr10_+_89622870 | 0.05 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr9_-_130712995 | 0.04 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr7_-_149470540 | 0.04 |
ENST00000302017.3
|
ZNF467
|
zinc finger protein 467 |
| chr6_+_26045603 | 0.04 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr15_+_52311398 | 0.04 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr22_+_24115000 | 0.04 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr13_+_49684445 | 0.04 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_+_52082751 | 0.04 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr1_-_152196669 | 0.04 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr12_-_109797249 | 0.04 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chrX_+_12993202 | 0.04 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr4_+_88896819 | 0.04 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_+_40915725 | 0.04 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr3_-_182698381 | 0.04 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr10_-_123274693 | 0.04 |
ENST00000429361.1
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr10_-_32217717 | 0.04 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr21_+_35553045 | 0.04 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_+_106067943 | 0.04 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr14_-_100772796 | 0.04 |
ENST00000554060.1
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr3_+_49840685 | 0.04 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr8_-_80993010 | 0.03 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr14_+_60712463 | 0.03 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_-_37899323 | 0.03 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_+_109577202 | 0.03 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr13_-_101327028 | 0.03 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr6_+_114178512 | 0.03 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr10_+_81466084 | 0.03 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr7_+_30068260 | 0.03 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr16_+_28875126 | 0.03 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr2_+_127413481 | 0.03 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr11_+_19798964 | 0.03 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr17_-_14683517 | 0.03 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr1_-_204436344 | 0.03 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr3_-_47023455 | 0.03 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr11_-_130184470 | 0.03 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr2_-_224810070 | 0.03 |
ENST00000429915.1
ENST00000233055.4 |
WDFY1
|
WD repeat and FYVE domain containing 1 |
| chr1_+_104198377 | 0.03 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr10_-_33247124 | 0.03 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr4_-_2264015 | 0.03 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr1_+_14026722 | 0.03 |
ENST00000376048.5
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr18_+_20715416 | 0.03 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr14_+_96829886 | 0.03 |
ENST00000556095.1
|
GSKIP
|
GSK3B interacting protein |
| chr3_+_182983090 | 0.03 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr11_+_64794991 | 0.03 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr5_+_149877440 | 0.03 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_-_17375541 | 0.03 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr5_-_146833803 | 0.03 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chrX_+_103173457 | 0.03 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr16_+_57680811 | 0.03 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr11_-_65793948 | 0.03 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr10_-_21806759 | 0.02 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr19_-_46145696 | 0.02 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr11_-_130184555 | 0.02 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr20_-_43150601 | 0.02 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr17_-_42441204 | 0.02 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr17_+_25621102 | 0.02 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr17_-_48207157 | 0.02 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr15_+_73976715 | 0.02 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr5_-_137071756 | 0.02 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr1_-_120190396 | 0.02 |
ENST00000421812.2
|
ZNF697
|
zinc finger protein 697 |
| chr11_-_104905840 | 0.02 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr8_+_133787586 | 0.02 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr12_-_105478339 | 0.02 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr16_+_57679859 | 0.02 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_-_79151695 | 0.02 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr16_+_57680840 | 0.02 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_104238912 | 0.02 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr7_+_69064300 | 0.02 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_33231268 | 0.02 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr14_-_96830207 | 0.02 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr1_+_93544821 | 0.02 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr18_+_74240756 | 0.02 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr5_-_146833485 | 0.02 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chrX_+_100645812 | 0.02 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr11_+_4116054 | 0.02 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr19_+_7580944 | 0.02 |
ENST00000597229.1
|
ZNF358
|
zinc finger protein 358 |
| chr2_-_122407007 | 0.02 |
ENST00000263710.4
ENST00000455322.2 ENST00000397587.3 ENST00000541377.1 |
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr11_+_4116005 | 0.02 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr2_+_187371440 | 0.02 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_+_99038919 | 0.02 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr17_-_53046058 | 0.02 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr12_-_15942309 | 0.02 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_+_228337079 | 0.02 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr3_-_105588231 | 0.02 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_+_26323764 | 0.02 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_225616515 | 0.02 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr16_+_28875268 | 0.02 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr13_+_32605437 | 0.02 |
ENST00000380250.3
|
FRY
|
furry homolog (Drosophila) |
| chr12_+_69633407 | 0.02 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr18_+_74240610 | 0.02 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr19_-_58514129 | 0.02 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr1_+_52521957 | 0.02 |
ENST00000472944.2
ENST00000484036.1 |
BTF3L4
|
basic transcription factor 3-like 4 |
| chr2_-_204399976 | 0.02 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr4_+_2819883 | 0.02 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr16_+_57679945 | 0.02 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr17_-_42100474 | 0.02 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr5_-_111754948 | 0.02 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr11_+_450255 | 0.02 |
ENST00000308020.5
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr16_+_57680043 | 0.02 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr10_+_69644404 | 0.01 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chrX_-_2418634 | 0.01 |
ENST00000444280.1
|
DHRSX
|
dehydrogenase/reductase (SDR family) X-linked |
| chr5_+_140593509 | 0.01 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr12_+_50898881 | 0.01 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr12_-_54778471 | 0.01 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr14_-_51297197 | 0.01 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr2_+_239756671 | 0.01 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr8_+_102504979 | 0.01 |
ENST00000395927.1
|
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chr5_+_149877334 | 0.01 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_-_149470297 | 0.01 |
ENST00000484747.1
|
ZNF467
|
zinc finger protein 467 |
| chr8_+_24151620 | 0.01 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr1_+_93544791 | 0.01 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr15_+_64388166 | 0.01 |
ENST00000353874.4
ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1
|
sorting nexin 1 |
| chr3_+_119187785 | 0.01 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chrX_-_38080077 | 0.01 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr7_-_128415844 | 0.01 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr14_-_100772767 | 0.01 |
ENST00000392908.3
ENST00000539621.1 |
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr17_+_67498538 | 0.01 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr10_+_123923205 | 0.01 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_-_498415 | 0.01 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr17_+_67498295 | 0.01 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_179050066 | 0.01 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr1_-_6546001 | 0.01 |
ENST00000400913.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr10_+_121578211 | 0.01 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr11_+_76494253 | 0.01 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr4_+_26322987 | 0.01 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_+_123923105 | 0.01 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr13_-_99630233 | 0.01 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr6_+_10556215 | 0.01 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr2_-_161350305 | 0.01 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chrX_+_100646190 | 0.01 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr12_-_25055177 | 0.01 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr13_-_24007815 | 0.01 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr1_+_112939121 | 0.01 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr2_-_122407097 | 0.01 |
ENST00000409078.3
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr22_-_19512893 | 0.01 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr2_-_9770706 | 0.01 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr1_-_2461684 | 0.01 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr9_+_128509624 | 0.01 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr14_-_100772862 | 0.01 |
ENST00000359232.3
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr9_-_107361788 | 0.01 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr16_+_54964740 | 0.01 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr17_-_62308087 | 0.01 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr21_+_33245548 | 0.01 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr17_-_33390667 | 0.01 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr11_+_111807863 | 0.01 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr2_-_157198860 | 0.01 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_65215604 | 0.01 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.0 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.0 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.0 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |