Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
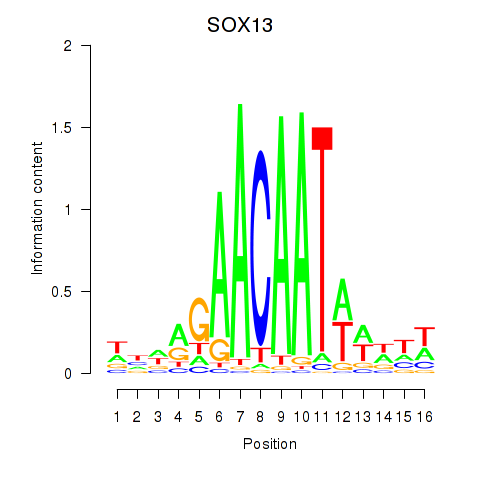
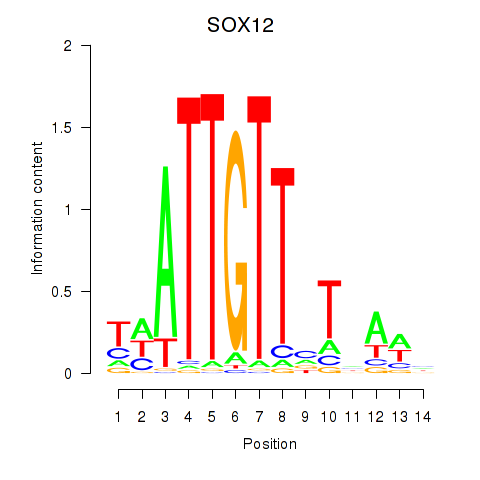
Results for SOX13_SOX12
Z-value: 1.07
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.10 | SRY-box transcription factor 13 |
|
SOX12
|
ENSG00000177732.6 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg19_v2_chr20_+_306177_306211 | 0.23 | 6.7e-01 | Click! |
| SOX13 | hg19_v2_chr1_+_204042723_204042784 | -0.01 | 9.8e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_136070614 | 0.99 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr14_+_39703112 | 0.87 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr19_+_21106081 | 0.85 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr12_-_10573149 | 0.82 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr4_-_74864386 | 0.82 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr11_-_85393886 | 0.76 |
ENST00000534224.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr11_-_8190534 | 0.70 |
ENST00000309737.6
ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3
|
RIC3 acetylcholine receptor chaperone |
| chr6_-_133055815 | 0.63 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr14_-_38036271 | 0.57 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr12_-_49581152 | 0.56 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr15_-_80263506 | 0.55 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr17_-_57229155 | 0.54 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chrX_+_107288280 | 0.53 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_153013588 | 0.53 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr12_+_11081828 | 0.50 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr1_+_153003671 | 0.48 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr11_-_18258342 | 0.47 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr17_-_38859996 | 0.45 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr1_-_153363452 | 0.45 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr1_-_152539248 | 0.44 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr1_-_153433120 | 0.44 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr2_+_132479948 | 0.42 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr20_-_52612705 | 0.42 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr8_+_24151620 | 0.40 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr6_-_133055896 | 0.40 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr20_-_52612468 | 0.40 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr12_-_10588539 | 0.40 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr3_+_98699880 | 0.37 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr19_-_21512144 | 0.36 |
ENST00000602023.1
|
ZNF708
|
zinc finger protein 708 |
| chr20_-_5426380 | 0.36 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr14_+_94577074 | 0.36 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr13_-_38172863 | 0.35 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr12_+_27623565 | 0.34 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr5_+_147582348 | 0.33 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr10_+_69865866 | 0.33 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr19_-_23433144 | 0.33 |
ENST00000418100.1
ENST00000597537.1 ENST00000597037.1 |
ZNF724P
|
zinc finger protein 724, pseudogene |
| chr2_-_25100893 | 0.33 |
ENST00000433852.1
|
ADCY3
|
adenylate cyclase 3 |
| chr6_+_12290586 | 0.33 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr4_-_112993808 | 0.33 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr3_-_178976996 | 0.33 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_11036844 | 0.32 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr8_-_54752406 | 0.32 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr6_-_134861089 | 0.32 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr6_+_76599809 | 0.31 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr18_+_74240610 | 0.31 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr19_-_21512202 | 0.31 |
ENST00000356929.3
|
ZNF708
|
zinc finger protein 708 |
| chr1_+_10057274 | 0.31 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr15_-_41166414 | 0.31 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr17_-_57158523 | 0.30 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr2_+_102927962 | 0.30 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr1_-_153029980 | 0.30 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr4_+_154622652 | 0.30 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr3_+_177534653 | 0.30 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr7_-_64023441 | 0.30 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr4_+_78829479 | 0.30 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr8_+_24151553 | 0.29 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_-_11175219 | 0.29 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr7_+_111846741 | 0.29 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr4_+_74301880 | 0.28 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr6_-_139613269 | 0.28 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr11_-_62609281 | 0.27 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr2_+_143635067 | 0.27 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr12_+_28605426 | 0.27 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr16_+_30709530 | 0.26 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr8_-_71157595 | 0.26 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr2_-_238499337 | 0.26 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr4_+_11470867 | 0.26 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr4_+_110749143 | 0.25 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr2_-_165630264 | 0.25 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr18_+_68002675 | 0.25 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr4_-_57547454 | 0.25 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr4_+_70916119 | 0.24 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr7_-_41742697 | 0.24 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr4_+_95128996 | 0.24 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_66317967 | 0.24 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr3_+_177545563 | 0.24 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr1_-_116383322 | 0.23 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr8_+_94752349 | 0.23 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr10_+_94594351 | 0.23 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr7_-_7575477 | 0.22 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr12_+_113354341 | 0.22 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_+_44809027 | 0.22 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr12_+_60058458 | 0.22 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_179318295 | 0.22 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr11_-_327537 | 0.22 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr10_+_118187379 | 0.21 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr7_+_871559 | 0.21 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr12_+_26205496 | 0.21 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr12_+_78224667 | 0.21 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr3_-_32544900 | 0.20 |
ENST00000205636.3
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr4_+_53728457 | 0.20 |
ENST00000248706.3
|
RASL11B
|
RAS-like, family 11, member B |
| chr8_+_105602961 | 0.20 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr21_+_43619796 | 0.20 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr12_-_76461249 | 0.20 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr6_-_108278456 | 0.20 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr21_+_17214724 | 0.20 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr7_+_7196565 | 0.20 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr4_+_187148556 | 0.20 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr6_-_26285737 | 0.20 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr3_+_119814070 | 0.20 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr12_-_68835991 | 0.20 |
ENST00000546086.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr7_-_35013217 | 0.20 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr13_-_49987885 | 0.19 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr4_+_95128748 | 0.19 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_+_64126503 | 0.19 |
ENST00000360117.3
|
ZNF107
|
zinc finger protein 107 |
| chr6_+_26217159 | 0.19 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr3_-_57678772 | 0.19 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr19_-_22018966 | 0.19 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr7_-_112635675 | 0.19 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr12_-_10601963 | 0.19 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr5_+_110073853 | 0.19 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr8_+_40010989 | 0.19 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr14_+_22600515 | 0.18 |
ENST00000390456.3
|
TRAV8-7
|
T cell receptor alpha variable 8-7 (non-functional) |
| chr16_+_89160217 | 0.18 |
ENST00000317447.4
ENST00000537290.1 |
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr11_-_85430088 | 0.18 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr8_-_141774467 | 0.18 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_212475148 | 0.18 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr17_-_71223839 | 0.18 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_-_135916073 | 0.18 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chrX_+_108780347 | 0.18 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_162101247 | 0.18 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr14_+_39944025 | 0.18 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr17_+_61151306 | 0.17 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr18_-_59415987 | 0.17 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr21_+_35553045 | 0.17 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr8_-_42358742 | 0.17 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr19_+_41882466 | 0.17 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr10_+_91461413 | 0.17 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr13_-_48575401 | 0.17 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr4_+_78804393 | 0.16 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chrX_-_19688475 | 0.16 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_+_58134756 | 0.16 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr5_+_35852797 | 0.16 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr4_-_122085469 | 0.16 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr12_+_28410128 | 0.16 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr19_+_35225060 | 0.16 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr11_+_59705928 | 0.16 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr19_+_36132631 | 0.16 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr4_+_75174180 | 0.16 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr6_+_126277842 | 0.16 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr6_-_26216872 | 0.15 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr2_+_109204909 | 0.15 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_+_26746951 | 0.15 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr17_-_72772462 | 0.15 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr5_-_86534822 | 0.15 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr11_+_128563652 | 0.15 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_79895097 | 0.15 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr8_-_61429315 | 0.15 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr6_-_26197478 | 0.15 |
ENST00000356476.2
|
HIST1H3D
|
histone cluster 1, H3d |
| chr13_-_25861530 | 0.15 |
ENST00000540661.1
|
MTMR6
|
myotubularin related protein 6 |
| chr8_+_86019382 | 0.14 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr16_-_11363178 | 0.14 |
ENST00000312693.3
|
TNP2
|
transition protein 2 (during histone to protamine replacement) |
| chr15_+_44119159 | 0.14 |
ENST00000263795.6
ENST00000381246.2 ENST00000452115.1 |
WDR76
|
WD repeat domain 76 |
| chr20_+_56964169 | 0.14 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr3_-_146262488 | 0.14 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr11_-_45928830 | 0.14 |
ENST00000449465.1
|
C11orf94
|
chromosome 11 open reading frame 94 |
| chr8_-_28965684 | 0.14 |
ENST00000523130.1
|
KIF13B
|
kinesin family member 13B |
| chr12_-_111142729 | 0.14 |
ENST00000546713.1
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr13_+_76413852 | 0.14 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr5_+_61874562 | 0.13 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr6_+_36839616 | 0.13 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr11_-_10879572 | 0.13 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr3_-_107777208 | 0.13 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr9_+_99690592 | 0.13 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr17_+_47448102 | 0.13 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr3_+_14693247 | 0.13 |
ENST00000383794.3
ENST00000303688.7 |
CCDC174
|
coiled-coil domain containing 174 |
| chr18_-_44500101 | 0.13 |
ENST00000589917.1
ENST00000587810.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr16_+_14802801 | 0.13 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr5_+_69321361 | 0.13 |
ENST00000515588.1
|
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr17_-_72772425 | 0.13 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr6_-_11382478 | 0.12 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr2_+_131769256 | 0.12 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr19_-_42192096 | 0.12 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr6_-_133035185 | 0.12 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr22_-_22292934 | 0.12 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr19_+_13858701 | 0.12 |
ENST00000586666.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr3_+_136649311 | 0.12 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr14_+_39703084 | 0.12 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr8_-_123139423 | 0.12 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr4_-_170679024 | 0.12 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr7_-_76247617 | 0.12 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_+_152975488 | 0.12 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr19_+_41281060 | 0.12 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr4_-_38858428 | 0.12 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr14_+_96949319 | 0.12 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr7_+_141811539 | 0.12 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr19_+_13842559 | 0.12 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr2_+_207220463 | 0.12 |
ENST00000598562.1
|
AC017081.1
|
Uncharacterized protein |
| chr5_+_81601166 | 0.12 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_+_26440700 | 0.12 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr1_+_207943667 | 0.11 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr11_-_72145426 | 0.11 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_-_156722195 | 0.11 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr13_+_111893533 | 0.11 |
ENST00000478679.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr14_+_38264418 | 0.11 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr2_+_232573208 | 0.11 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr10_+_81462983 | 0.11 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr19_-_42192189 | 0.11 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr10_+_27793197 | 0.11 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr17_-_37764128 | 0.11 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.3 | GO:0060585 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.4 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.2 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.3 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 1.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.0 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.0 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.0 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 1.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.0 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |