Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
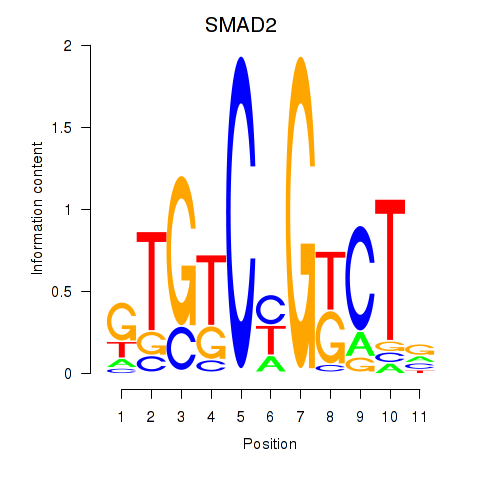
Results for SMAD2
Z-value: 0.43
Transcription factors associated with SMAD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD2
|
ENSG00000175387.11 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD2 | hg19_v2_chr18_-_45456693_45456733 | 0.77 | 7.4e-02 | Click! |
Activity profile of SMAD2 motif
Sorted Z-values of SMAD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22676808 | 0.30 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr8_-_123706338 | 0.22 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr7_+_150758642 | 0.18 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr3_+_194406603 | 0.17 |
ENST00000329759.4
|
FAM43A
|
family with sequence similarity 43, member A |
| chr6_-_134861089 | 0.17 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr10_+_72972281 | 0.13 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr7_-_96654133 | 0.13 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr12_+_53443963 | 0.13 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr6_+_32132360 | 0.12 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr1_-_152539248 | 0.11 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr12_-_6740802 | 0.11 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr16_-_425205 | 0.10 |
ENST00000448854.1
|
TMEM8A
|
transmembrane protein 8A |
| chr3_-_88108192 | 0.10 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr20_+_44098346 | 0.10 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr16_-_70719925 | 0.10 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr6_-_31612808 | 0.10 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_102158157 | 0.10 |
ENST00000541662.1
ENST00000306682.6 ENST00000465829.1 |
RASA4B
|
RAS p21 protein activator 4B |
| chr7_-_102257139 | 0.10 |
ENST00000521076.1
ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4
|
RAS p21 protein activator 4 |
| chr8_-_130587237 | 0.09 |
ENST00000520048.1
|
CCDC26
|
coiled-coil domain containing 26 |
| chr11_-_118789613 | 0.09 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr4_-_5890145 | 0.08 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr19_-_55580838 | 0.08 |
ENST00000396247.3
ENST00000586945.1 ENST00000587026.1 |
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr17_-_58469591 | 0.08 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr12_-_108991778 | 0.08 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr22_+_22764088 | 0.08 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chrX_+_48660565 | 0.08 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr16_+_2198604 | 0.07 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr21_-_36259445 | 0.07 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr6_-_143771799 | 0.07 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chr5_-_151066514 | 0.07 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr1_-_207119738 | 0.07 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr1_+_33722080 | 0.07 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr19_+_49128209 | 0.07 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr16_+_31483374 | 0.07 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr3_+_195447738 | 0.07 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr12_+_53443680 | 0.07 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr21_+_47401650 | 0.07 |
ENST00000361866.3
|
COL6A1
|
collagen, type VI, alpha 1 |
| chr3_-_88108212 | 0.06 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr4_+_102268904 | 0.06 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr8_-_96281419 | 0.06 |
ENST00000286688.5
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr16_+_24621546 | 0.06 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr15_+_74509530 | 0.06 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr1_-_44497118 | 0.06 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_130829588 | 0.06 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr4_-_114682719 | 0.06 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr10_+_102729249 | 0.06 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr7_-_130080681 | 0.05 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr11_-_74204742 | 0.05 |
ENST00000310109.4
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr20_+_44098385 | 0.05 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr16_-_3355645 | 0.05 |
ENST00000396862.1
ENST00000573608.1 |
TIGD7
|
tigger transposable element derived 7 |
| chr14_+_36295638 | 0.05 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chrX_-_107018969 | 0.05 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr20_+_34680620 | 0.05 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr3_-_195163584 | 0.05 |
ENST00000439666.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr11_-_117166201 | 0.05 |
ENST00000510915.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr6_-_159466042 | 0.05 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr4_-_103749205 | 0.05 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr22_-_43567750 | 0.05 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr9_+_140145713 | 0.05 |
ENST00000388931.3
ENST00000412566.1 |
C9orf173
|
chromosome 9 open reading frame 173 |
| chr1_+_174933899 | 0.05 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_+_36295504 | 0.05 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr12_+_68042495 | 0.05 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr20_+_34020827 | 0.04 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr15_+_45879964 | 0.04 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr10_-_75193308 | 0.04 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr4_-_103749313 | 0.04 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_51937331 | 0.04 |
ENST00000310914.5
|
IQCF1
|
IQ motif containing F1 |
| chr22_+_45072958 | 0.04 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr20_+_43343517 | 0.04 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr12_-_22094336 | 0.04 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr3_+_3841108 | 0.04 |
ENST00000319331.3
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chrX_-_48328631 | 0.04 |
ENST00000429543.1
ENST00000317669.5 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr12_+_93965609 | 0.04 |
ENST00000549887.1
ENST00000551556.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr2_-_220173685 | 0.04 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr6_-_152489484 | 0.04 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chrX_-_47489244 | 0.04 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr12_-_120241187 | 0.04 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr19_-_14606900 | 0.04 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr2_-_72375167 | 0.04 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr12_+_104982622 | 0.04 |
ENST00000549016.1
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr15_+_75498739 | 0.04 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr10_-_14372870 | 0.04 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr2_-_17981462 | 0.04 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr19_+_56186557 | 0.04 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr5_+_139493665 | 0.04 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr2_-_220174166 | 0.03 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr8_-_91657740 | 0.03 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr19_-_46272462 | 0.03 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr4_-_102268708 | 0.03 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr5_+_133861339 | 0.03 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr6_-_159466136 | 0.03 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr11_+_64808675 | 0.03 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr4_-_103749105 | 0.03 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_+_74204395 | 0.03 |
ENST00000526036.1
|
AP001372.2
|
AP001372.2 |
| chr22_+_19702069 | 0.03 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr1_+_2407754 | 0.03 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr12_-_57472522 | 0.03 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr4_-_39979576 | 0.03 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr11_-_22647350 | 0.03 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr3_+_123813509 | 0.03 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr8_-_145641864 | 0.03 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr12_-_47473557 | 0.03 |
ENST00000321382.3
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr13_+_25670268 | 0.03 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr11_+_75273101 | 0.03 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr9_-_136006496 | 0.03 |
ENST00000372062.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr6_-_30523865 | 0.02 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chrX_-_107019181 | 0.02 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr9_-_34523027 | 0.02 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr3_+_150321068 | 0.02 |
ENST00000471696.1
ENST00000477889.1 ENST00000485923.1 |
SELT
|
Selenoprotein T |
| chrX_+_153029633 | 0.02 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr1_+_16693578 | 0.02 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr6_+_147525541 | 0.02 |
ENST00000367481.3
ENST00000546097.1 |
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr12_+_101188547 | 0.02 |
ENST00000546991.1
ENST00000392979.3 |
ANO4
|
anoctamin 4 |
| chr11_-_33913708 | 0.02 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr16_+_58059470 | 0.02 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr1_-_201915590 | 0.02 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr5_+_64064748 | 0.02 |
ENST00000381070.3
ENST00000508024.1 |
CWC27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_-_184243561 | 0.02 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr22_+_45072925 | 0.02 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr15_+_68570062 | 0.02 |
ENST00000306917.4
|
FEM1B
|
fem-1 homolog b (C. elegans) |
| chr11_-_3859089 | 0.02 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr3_+_88108381 | 0.02 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr15_-_82338460 | 0.02 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr1_-_149908217 | 0.02 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chrX_-_48328551 | 0.02 |
ENST00000376876.3
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr12_-_22094159 | 0.02 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_-_3571934 | 0.02 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr20_+_36888551 | 0.02 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr13_-_80915059 | 0.02 |
ENST00000377104.3
|
SPRY2
|
sprouty homolog 2 (Drosophila) |
| chr1_-_1243252 | 0.02 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr12_-_47473642 | 0.02 |
ENST00000266581.4
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr1_+_93913665 | 0.02 |
ENST00000271234.7
ENST00000370256.4 ENST00000260506.8 |
FNBP1L
|
formin binding protein 1-like |
| chr19_+_56186606 | 0.02 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
| chr14_+_62162258 | 0.02 |
ENST00000337138.4
ENST00000394997.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr7_-_559853 | 0.02 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr11_+_7597639 | 0.02 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_+_50094866 | 0.02 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr11_-_65149422 | 0.02 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr4_-_103749179 | 0.02 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_41707778 | 0.02 |
ENST00000337495.5
ENST00000372597.1 ENST00000372596.1 |
SCMH1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr9_-_73029540 | 0.02 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chrX_+_38420783 | 0.02 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr1_+_15802594 | 0.01 |
ENST00000375910.3
|
CELA2B
|
chymotrypsin-like elastase family, member 2B |
| chr3_-_79068138 | 0.01 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_46288917 | 0.01 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr1_+_28052518 | 0.01 |
ENST00000530324.1
ENST00000234549.7 ENST00000373949.1 ENST00000010299.6 |
FAM76A
|
family with sequence similarity 76, member A |
| chr5_-_64064508 | 0.01 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr19_-_46105411 | 0.01 |
ENST00000323040.4
ENST00000544371.1 |
GPR4
OPA3
|
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr10_-_102046417 | 0.01 |
ENST00000370372.2
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr17_+_73629500 | 0.01 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr1_+_28052456 | 0.01 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr12_+_7037461 | 0.01 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr1_+_100111479 | 0.01 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr19_+_11466167 | 0.01 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr1_+_26872324 | 0.01 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr16_+_57702099 | 0.01 |
ENST00000333493.4
ENST00000327655.6 |
GPR97
|
G protein-coupled receptor 97 |
| chr17_-_46623441 | 0.01 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr8_+_95907993 | 0.01 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_-_47473425 | 0.01 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr13_+_96743093 | 0.01 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr9_+_71944241 | 0.01 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr20_+_43343476 | 0.01 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr10_-_102046098 | 0.01 |
ENST00000441611.1
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr20_-_48532046 | 0.01 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr17_-_15165854 | 0.01 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
| chr16_-_4987065 | 0.01 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chrY_+_16634483 | 0.01 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr19_+_13106383 | 0.01 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_-_71003568 | 0.01 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_130569592 | 0.01 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_130569429 | 0.01 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_-_130080977 | 0.01 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr3_-_195603566 | 0.01 |
ENST00000424563.1
ENST00000411741.1 |
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chr22_+_38035623 | 0.01 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr12_+_67663056 | 0.01 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr3_-_195163803 | 0.01 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr1_-_935491 | 0.01 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr5_+_140345820 | 0.01 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr5_-_150521192 | 0.01 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr14_+_23025534 | 0.01 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr20_-_45976816 | 0.01 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_122516626 | 0.01 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr16_-_54320675 | 0.01 |
ENST00000329734.3
|
IRX3
|
iroquois homeobox 3 |
| chr1_-_160040038 | 0.01 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chrX_-_99987088 | 0.01 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr19_-_52150053 | 0.01 |
ENST00000599649.1
ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5
SIGLEC14
SIGLEC5
|
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr3_-_150320937 | 0.01 |
ENST00000479209.1
|
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr6_+_30853002 | 0.01 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr16_+_30406423 | 0.01 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr20_+_43343886 | 0.01 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr13_-_88323514 | 0.01 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr13_-_96705624 | 0.01 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr1_+_100111580 | 0.01 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr12_+_68042517 | 0.01 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr17_+_40440094 | 0.01 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chrX_+_24167746 | 0.01 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr8_-_102217796 | 0.01 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr3_+_123813543 | 0.01 |
ENST00000360013.3
|
KALRN
|
kalirin, RhoGEF kinase |
| chr16_-_11681316 | 0.01 |
ENST00000571688.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr8_+_144373550 | 0.00 |
ENST00000330143.3
ENST00000521537.1 ENST00000518432.1 ENST00000520333.1 |
ZNF696
|
zinc finger protein 696 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.0 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |