Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
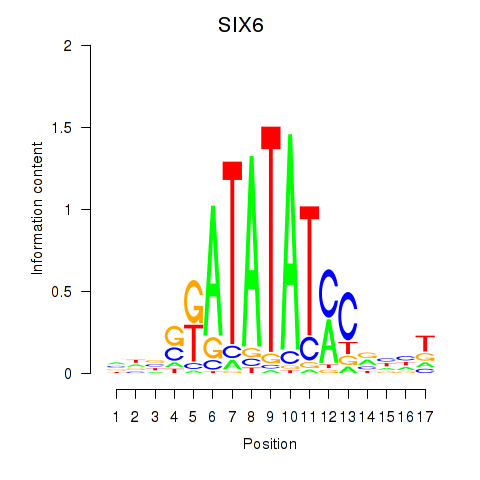
Results for SIX6
Z-value: 0.45
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_71104588 | 0.62 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr3_+_186742464 | 0.47 |
ENST00000416235.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr8_-_82598067 | 0.42 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr9_+_93759317 | 0.41 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr11_-_104916034 | 0.20 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr11_+_74699703 | 0.18 |
ENST00000529024.1
ENST00000544263.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr1_+_160709076 | 0.18 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_160709029 | 0.17 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_40942887 | 0.16 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr1_+_160709055 | 0.15 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr10_+_91061712 | 0.15 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr1_-_67142710 | 0.13 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr12_+_122018697 | 0.12 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr5_+_55147205 | 0.11 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr3_+_8543561 | 0.11 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr12_+_133707570 | 0.11 |
ENST00000416488.1
ENST00000540096.2 |
ZNF268
CTD-2140B24.4
|
zinc finger protein 268 Zinc finger protein 268 |
| chr11_-_104972158 | 0.11 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr2_-_134326009 | 0.10 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr16_+_22524379 | 0.10 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr9_-_21142144 | 0.10 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr12_-_14849470 | 0.10 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr20_+_54987305 | 0.09 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_+_179263308 | 0.09 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chrX_+_12809463 | 0.09 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr8_+_1993173 | 0.09 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr18_-_69246167 | 0.09 |
ENST00000566582.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr20_+_54987168 | 0.08 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr5_-_78281775 | 0.08 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr16_+_28648975 | 0.08 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr22_+_20877924 | 0.07 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr2_+_27760247 | 0.07 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr14_-_46185155 | 0.07 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr10_+_81891416 | 0.07 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr15_-_51397473 | 0.07 |
ENST00000327536.5
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr7_-_123198284 | 0.06 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr14_-_31926595 | 0.06 |
ENST00000547378.1
|
RP11-176H8.1
|
Uncharacterized protein |
| chr1_-_150669604 | 0.06 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr16_+_19467772 | 0.06 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr7_-_81635106 | 0.06 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_+_40787194 | 0.06 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr9_-_135230336 | 0.06 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr8_+_1993152 | 0.05 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr9_+_109685630 | 0.05 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr6_+_37012607 | 0.05 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr20_+_34802295 | 0.05 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr12_+_119616447 | 0.05 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr18_-_69246186 | 0.05 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr12_+_19282643 | 0.05 |
ENST00000317589.4
ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr8_+_32579321 | 0.05 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chrX_-_18238999 | 0.05 |
ENST00000380033.4
ENST00000380030.3 |
BEND2
|
BEN domain containing 2 |
| chr19_-_16008880 | 0.05 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr5_+_115420688 | 0.05 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr11_+_122709200 | 0.05 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr6_+_80816342 | 0.05 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr19_+_31658405 | 0.05 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr2_-_228497888 | 0.04 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr19_+_46000479 | 0.04 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr4_+_113739244 | 0.04 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr9_-_23821842 | 0.04 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr8_-_124553437 | 0.04 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr5_+_140201183 | 0.04 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr10_-_61720640 | 0.04 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr11_+_34999328 | 0.04 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr12_-_46385811 | 0.04 |
ENST00000419565.2
|
SCAF11
|
SR-related CTD-associated factor 11 |
| chr2_-_71222466 | 0.03 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr1_-_182369751 | 0.03 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr10_-_79397479 | 0.03 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_121476750 | 0.03 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr15_+_63414760 | 0.03 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr14_+_21525981 | 0.03 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr3_-_9994021 | 0.03 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr1_+_212738676 | 0.03 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr9_-_67786624 | 0.03 |
ENST00000455764.2
|
FAM27E3
|
family with sequence similarity 27, member E3 |
| chr17_-_5342380 | 0.03 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr14_+_21510385 | 0.03 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr2_+_62132781 | 0.03 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr1_+_156119466 | 0.03 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr22_-_17702729 | 0.02 |
ENST00000449907.2
ENST00000441548.1 ENST00000399839.1 |
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr19_-_14628645 | 0.02 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr18_+_158327 | 0.02 |
ENST00000582707.1
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr3_+_125694347 | 0.02 |
ENST00000505382.1
ENST00000511082.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr1_-_157014865 | 0.02 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr1_-_10003372 | 0.02 |
ENST00000377223.1
ENST00000541052.1 ENST00000377213.1 |
LZIC
|
leucine zipper and CTNNBIP1 domain containing |
| chr3_+_8543393 | 0.02 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_-_36235559 | 0.02 |
ENST00000251195.5
|
CLSPN
|
claspin |
| chr11_+_74699942 | 0.02 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr20_-_4990931 | 0.02 |
ENST00000379333.1
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr3_-_196242233 | 0.02 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chrX_+_8433376 | 0.02 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr12_-_11091862 | 0.02 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr12_-_121477039 | 0.02 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr9_-_99382065 | 0.02 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr6_+_127898312 | 0.02 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr5_+_40909354 | 0.02 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr10_-_14614311 | 0.02 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr2_-_62081148 | 0.01 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr12_+_133614062 | 0.01 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr11_+_112041253 | 0.01 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr10_-_75008620 | 0.01 |
ENST00000372950.4
|
DNAJC9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr12_-_498620 | 0.01 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr2_+_177134134 | 0.01 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr12_-_10601963 | 0.01 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr15_+_58430567 | 0.01 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr16_-_21436459 | 0.01 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr7_-_44530479 | 0.01 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr8_+_26247878 | 0.01 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr5_-_117897751 | 0.00 |
ENST00000515704.1
ENST00000506769.1 |
CTD-2281M20.1
|
CTD-2281M20.1 |
| chr16_+_66410244 | 0.00 |
ENST00000562048.1
ENST00000568155.2 |
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr13_-_46679185 | 0.00 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_45307817 | 0.00 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr22_+_18721427 | 0.00 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr17_-_6524159 | 0.00 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr15_+_58430368 | 0.00 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr8_+_32579341 | 0.00 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chrX_+_128674213 | 0.00 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chr1_-_161015663 | 0.00 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr4_+_189321881 | 0.00 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.0 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |