Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
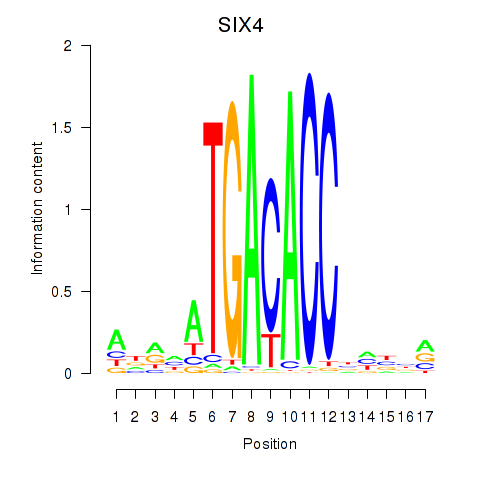
Results for SIX4
Z-value: 0.46
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.8 | SIX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX4 | hg19_v2_chr14_-_61191049_61191080 | 0.32 | 5.3e-01 | Click! |
Activity profile of SIX4 motif
Sorted Z-values of SIX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_123139423 | 0.42 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr20_-_45530365 | 0.33 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr8_-_6115044 | 0.31 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr16_+_640201 | 0.29 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr12_+_30948600 | 0.28 |
ENST00000550292.1
|
LINC00941
|
long intergenic non-protein coding RNA 941 |
| chr14_+_38065052 | 0.24 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr17_-_191188 | 0.18 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr3_-_93692681 | 0.15 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr12_-_10282742 | 0.13 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr5_+_78365536 | 0.13 |
ENST00000255192.3
|
BHMT2
|
betaine--homocysteine S-methyltransferase 2 |
| chr1_+_35734616 | 0.12 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr2_+_85811525 | 0.10 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr4_+_11470867 | 0.10 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr1_-_32264356 | 0.10 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr1_+_149239529 | 0.10 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr11_-_96076334 | 0.08 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr1_-_109849612 | 0.08 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chr11_-_18956556 | 0.07 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr19_+_13135439 | 0.07 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr10_+_71444655 | 0.07 |
ENST00000434931.2
|
RP11-242G20.1
|
Uncharacterized protein |
| chr7_-_111428957 | 0.07 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr9_-_95298314 | 0.07 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr7_+_69064300 | 0.07 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr12_+_43086018 | 0.07 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr10_+_7860460 | 0.07 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr19_+_13135386 | 0.07 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_152196669 | 0.06 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr2_+_219135115 | 0.06 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr12_+_133757995 | 0.06 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr1_+_8378140 | 0.05 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr3_+_32147997 | 0.05 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr22_-_39268192 | 0.05 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr2_-_74710078 | 0.05 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr8_+_132916318 | 0.05 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr19_+_13134772 | 0.05 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_+_128458514 | 0.05 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr20_+_816695 | 0.05 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr7_+_2687173 | 0.05 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr2_-_169104651 | 0.05 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr11_-_35547572 | 0.05 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr1_+_38022572 | 0.05 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr22_-_39268308 | 0.04 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr9_-_95298254 | 0.04 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr3_-_49158218 | 0.04 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr12_-_58135903 | 0.04 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_-_183967296 | 0.04 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr6_-_27840099 | 0.04 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr11_+_117049445 | 0.04 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_-_130184470 | 0.04 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr17_-_39684550 | 0.04 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr1_+_26737292 | 0.04 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr15_+_41245160 | 0.04 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr11_+_117947724 | 0.04 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr19_+_50887585 | 0.04 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr13_+_33160553 | 0.04 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr14_-_67981916 | 0.04 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr5_+_179135246 | 0.03 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr11_-_113644491 | 0.03 |
ENST00000200135.3
|
ZW10
|
zw10 kinetochore protein |
| chr11_+_117049854 | 0.03 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr1_+_156611704 | 0.03 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr5_-_78365437 | 0.03 |
ENST00000380311.4
ENST00000540686.1 ENST00000255189.3 |
DMGDH
|
dimethylglycine dehydrogenase |
| chr11_+_117947782 | 0.03 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr14_-_67981870 | 0.03 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_-_61523622 | 0.02 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr14_-_106692191 | 0.02 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr10_+_89622870 | 0.02 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr5_+_78365577 | 0.02 |
ENST00000518666.1
ENST00000521567.1 |
BHMT2
|
betaine--homocysteine S-methyltransferase 2 |
| chr1_+_116184566 | 0.02 |
ENST00000355485.2
ENST00000369510.4 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chr2_+_120687335 | 0.02 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr19_+_30414551 | 0.02 |
ENST00000360605.4
ENST00000570564.1 ENST00000574233.1 ENST00000585655.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr11_+_2415061 | 0.02 |
ENST00000481687.1
|
CD81
|
CD81 molecule |
| chr11_+_117049910 | 0.02 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr5_+_85913721 | 0.02 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr17_-_72527605 | 0.02 |
ENST00000392621.1
ENST00000314401.3 |
CD300LB
|
CD300 molecule-like family member b |
| chr14_-_92413353 | 0.02 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr19_+_50270219 | 0.01 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr4_-_2420357 | 0.01 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chrX_-_119763835 | 0.01 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr7_-_26578407 | 0.01 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr3_-_183966717 | 0.01 |
ENST00000446569.1
ENST00000418734.2 ENST00000397676.3 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr11_+_33279850 | 0.01 |
ENST00000531504.1
ENST00000456517.1 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr20_-_52687059 | 0.01 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr11_-_130184555 | 0.01 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr20_-_43150601 | 0.01 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr2_-_180427304 | 0.01 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr1_+_231473743 | 0.01 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr1_-_247335269 | 0.01 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr11_+_34073195 | 0.01 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr11_-_64511575 | 0.01 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_2906979 | 0.01 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr5_+_149569520 | 0.01 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr3_-_69249863 | 0.01 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr19_+_11466167 | 0.00 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr19_+_19174795 | 0.00 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr2_-_61244308 | 0.00 |
ENST00000407787.1
ENST00000398658.2 |
PUS10
|
pseudouridylate synthase 10 |
| chr17_+_7465216 | 0.00 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr1_+_116915855 | 0.00 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr7_+_97361388 | 0.00 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.2 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |