Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
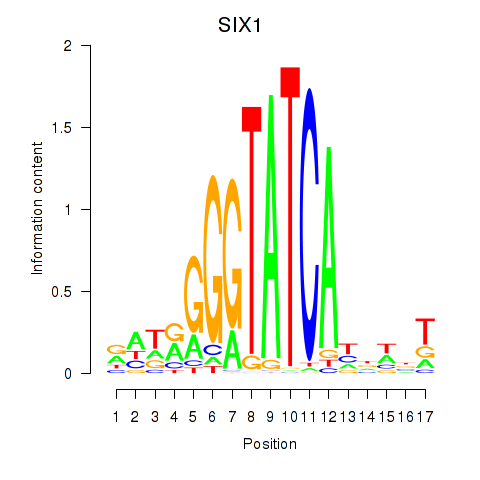
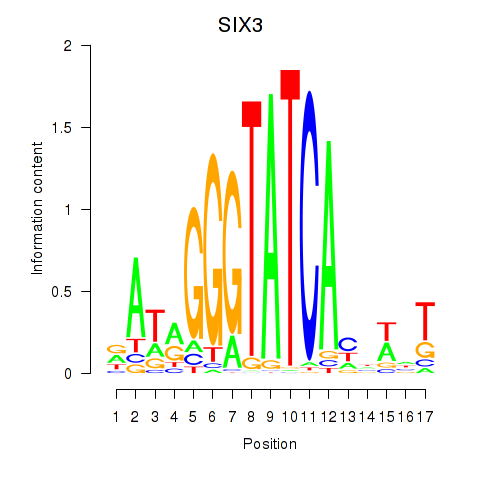
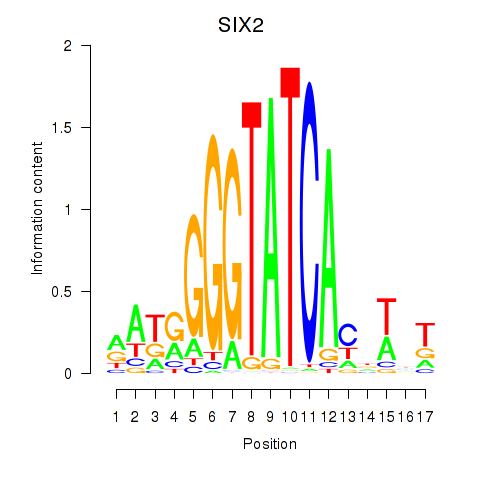
Results for SIX1_SIX3_SIX2
Z-value: 0.33
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.47 | 3.5e-01 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.40 | 4.3e-01 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | -0.12 | 8.2e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_171640291 | 0.23 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr14_+_51026844 | 0.13 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr6_+_53794780 | 0.12 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr3_+_184530173 | 0.11 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_169337176 | 0.11 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr7_+_99746514 | 0.11 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr19_+_42824511 | 0.10 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chr16_+_22517166 | 0.10 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr3_+_42850959 | 0.09 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr1_+_152730499 | 0.08 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr9_-_113018746 | 0.08 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr11_-_118134997 | 0.08 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr16_-_28550320 | 0.08 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr1_+_40942887 | 0.08 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr1_-_51791596 | 0.08 |
ENST00000532836.1
ENST00000422925.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr3_+_52448539 | 0.07 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr19_-_43702231 | 0.07 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr8_-_123139423 | 0.07 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr20_-_55100981 | 0.07 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr19_+_50015870 | 0.07 |
ENST00000599701.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_11685345 | 0.07 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr19_+_42055879 | 0.07 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr3_+_178865887 | 0.07 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr11_-_59633951 | 0.07 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr19_-_42916499 | 0.07 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr10_-_38265517 | 0.06 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr5_-_142077569 | 0.06 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr1_-_207119738 | 0.06 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr19_+_17579556 | 0.06 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr1_+_241695424 | 0.06 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_105953912 | 0.05 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr16_-_28550348 | 0.05 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr8_-_95487272 | 0.05 |
ENST00000297592.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr12_-_7077125 | 0.05 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr9_-_27005686 | 0.05 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chrX_+_13587712 | 0.05 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr15_-_100258029 | 0.05 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr4_+_70894130 | 0.05 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_-_171711387 | 0.05 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr7_+_75511362 | 0.05 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr1_+_160709029 | 0.05 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr14_+_62462541 | 0.05 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chrX_+_1733876 | 0.04 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr14_+_24025345 | 0.04 |
ENST00000557630.1
|
THTPA
|
thiamine triphosphatase |
| chr1_+_87595433 | 0.04 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr11_-_82782952 | 0.04 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr7_+_138915102 | 0.04 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr1_+_241695670 | 0.04 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_114514181 | 0.04 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr4_-_156298087 | 0.04 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr4_+_11470867 | 0.04 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr10_+_18948311 | 0.04 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr19_-_37407172 | 0.04 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr11_+_17298522 | 0.04 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr4_-_156297949 | 0.04 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr4_+_78829479 | 0.04 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr19_+_11039391 | 0.04 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr6_+_53964336 | 0.04 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chr15_-_31521567 | 0.04 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr7_-_92219698 | 0.04 |
ENST00000438306.1
ENST00000445716.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr12_+_2912363 | 0.04 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr15_+_91416092 | 0.04 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr16_+_28763108 | 0.04 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr1_+_198189921 | 0.04 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr9_+_67977438 | 0.04 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr7_-_34978980 | 0.04 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr8_+_121457642 | 0.04 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr11_-_76155700 | 0.04 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr6_+_96969672 | 0.04 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr2_-_150444116 | 0.04 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr3_-_125900369 | 0.04 |
ENST00000490367.1
|
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr3_-_178976996 | 0.04 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr11_+_100862811 | 0.04 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr5_+_71616188 | 0.04 |
ENST00000380639.5
ENST00000543322.1 ENST00000503868.1 ENST00000510676.2 ENST00000536805.1 |
PTCD2
|
pentatricopeptide repeat domain 2 |
| chr14_+_61449076 | 0.04 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr1_+_214776516 | 0.04 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr20_+_36661910 | 0.04 |
ENST00000373433.4
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr12_+_7052974 | 0.04 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr16_+_22524379 | 0.04 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr22_+_37257015 | 0.04 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr4_+_129349188 | 0.04 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr10_-_103603523 | 0.04 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr2_+_74361599 | 0.03 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr18_+_61575200 | 0.03 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr11_+_17298543 | 0.03 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr16_+_451826 | 0.03 |
ENST00000219481.5
ENST00000397710.1 ENST00000424398.2 |
DECR2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr12_+_147052 | 0.03 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr20_+_62694834 | 0.03 |
ENST00000415602.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr19_-_52097613 | 0.03 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr3_+_113609472 | 0.03 |
ENST00000472026.1
ENST00000462838.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr4_-_156298028 | 0.03 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr2_-_55459294 | 0.03 |
ENST00000407122.1
ENST00000406437.2 |
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr11_+_85339623 | 0.03 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr7_-_92219337 | 0.03 |
ENST00000456502.1
ENST00000427372.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr18_-_51750948 | 0.03 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr11_+_27062502 | 0.03 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_159846066 | 0.03 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr13_-_60737898 | 0.03 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr10_+_48359344 | 0.03 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr10_-_33405600 | 0.03 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr13_-_41837620 | 0.03 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr10_+_51549498 | 0.03 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_-_108743471 | 0.03 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_-_149095652 | 0.03 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr1_+_16330723 | 0.03 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
| chr6_+_31554962 | 0.03 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr19_+_11457232 | 0.03 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr19_-_36606181 | 0.03 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr9_+_97766409 | 0.03 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr11_+_114168085 | 0.03 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr11_-_112034831 | 0.03 |
ENST00000280357.7
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr9_+_12693336 | 0.03 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr2_+_161993465 | 0.03 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_5238793 | 0.03 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr16_-_47493041 | 0.03 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr20_+_44074071 | 0.03 |
ENST00000595203.1
|
AL031663.2
|
CDNA FLJ26875 fis, clone PRS08969; Uncharacterized protein |
| chr11_-_35547151 | 0.03 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr12_+_7053172 | 0.03 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr3_-_178984759 | 0.03 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_-_46786245 | 0.03 |
ENST00000442359.2
|
PRSS45
|
protease, serine, 45 |
| chr12_+_49717081 | 0.03 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr2_-_207629997 | 0.03 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr22_-_44258360 | 0.03 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr3_+_141144963 | 0.03 |
ENST00000510726.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_120517174 | 0.03 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr9_+_97766469 | 0.03 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr4_-_14889791 | 0.03 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr7_+_12727250 | 0.03 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr17_+_43213004 | 0.02 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr6_-_138428613 | 0.02 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr1_+_160709076 | 0.02 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr4_-_153332886 | 0.02 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_-_141392538 | 0.02 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr16_+_27214802 | 0.02 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr3_-_10028366 | 0.02 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr21_+_37442239 | 0.02 |
ENST00000530908.1
ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr1_+_53793885 | 0.02 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr8_-_48651648 | 0.02 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr2_-_152146385 | 0.02 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr22_-_43091639 | 0.02 |
ENST00000249005.2
ENST00000381278.3 |
A4GALT
|
alpha 1,4-galactosyltransferase |
| chr12_-_112614506 | 0.02 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr2_-_180871780 | 0.02 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_-_52883786 | 0.02 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr15_+_62853562 | 0.02 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr14_+_73563735 | 0.02 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr5_-_143550241 | 0.02 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr14_-_94759595 | 0.02 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr4_+_159131346 | 0.02 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr11_-_108438847 | 0.02 |
ENST00000531386.1
|
EXPH5
|
exophilin 5 |
| chr19_+_37464063 | 0.02 |
ENST00000586353.1
ENST00000433993.1 ENST00000592567.1 |
ZNF568
|
zinc finger protein 568 |
| chr10_-_103603568 | 0.02 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_-_11036844 | 0.02 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr12_-_58135903 | 0.02 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_68279256 | 0.02 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr11_-_111794446 | 0.02 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr1_+_87595497 | 0.02 |
ENST00000471417.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr2_-_207630033 | 0.02 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr1_+_54703710 | 0.02 |
ENST00000361350.4
|
SSBP3-AS1
|
SSBP3 antisense RNA 1 |
| chr6_-_35656685 | 0.02 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr1_+_174670143 | 0.02 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr21_+_35736302 | 0.02 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr3_-_155011483 | 0.02 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr4_-_860950 | 0.02 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr2_-_150444300 | 0.02 |
ENST00000303319.5
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr5_+_81575281 | 0.02 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_-_11779174 | 0.02 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_-_50836762 | 0.02 |
ENST00000474951.1
ENST00000391818.2 |
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| chr6_-_35656712 | 0.02 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr19_-_11457162 | 0.02 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr13_+_28712614 | 0.02 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr7_-_123198284 | 0.02 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr12_+_104680793 | 0.02 |
ENST00000529546.1
ENST00000529751.1 ENST00000540716.1 ENST00000528079.2 ENST00000526580.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr12_-_118796971 | 0.02 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr3_+_184529948 | 0.02 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr2_-_158732340 | 0.02 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr5_-_133304473 | 0.02 |
ENST00000231512.3
|
C5orf15
|
chromosome 5 open reading frame 15 |
| chr19_+_47523058 | 0.02 |
ENST00000602212.1
ENST00000602189.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr19_+_36632204 | 0.02 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr22_+_21321531 | 0.02 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr19_+_11457175 | 0.02 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr12_+_16500037 | 0.02 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr12_-_110883346 | 0.02 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_+_933555 | 0.02 |
ENST00000534485.1
|
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr1_+_26737292 | 0.02 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr2_+_61404624 | 0.02 |
ENST00000394457.3
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr19_-_3600549 | 0.02 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr1_-_44818599 | 0.02 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr4_+_26165074 | 0.02 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_+_152907913 | 0.02 |
ENST00000370167.4
|
DUSP9
|
dual specificity phosphatase 9 |
| chr12_-_323689 | 0.02 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr16_+_2303738 | 0.02 |
ENST00000454671.1
|
AC009065.1
|
Uncharacterized protein |
| chr6_-_159466042 | 0.02 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr5_+_140201183 | 0.02 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr15_+_89631381 | 0.02 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr10_-_96122682 | 0.02 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr11_+_58874701 | 0.02 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr16_+_11343475 | 0.02 |
ENST00000572173.1
|
RMI2
|
RecQ mediated genome instability 2 |
| chr12_+_26205496 | 0.02 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr10_+_104613980 | 0.02 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |