Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
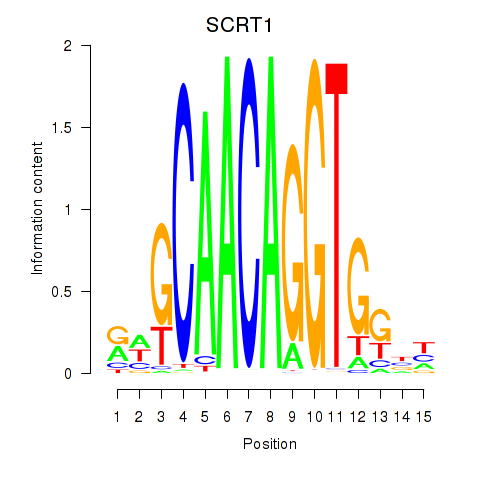
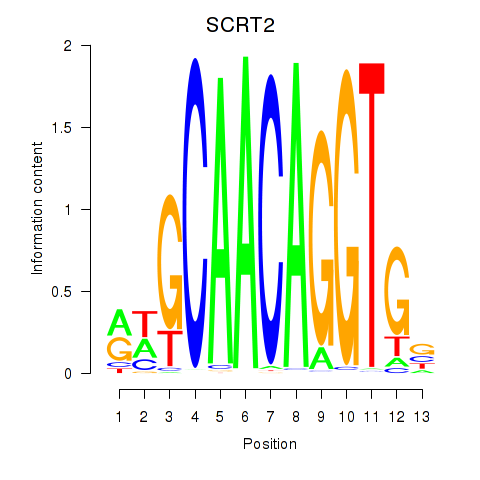
Results for SCRT1_SCRT2
Z-value: 1.14
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.16 | 7.6e-01 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_165675197 | 1.40 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr4_-_6675550 | 0.85 |
ENST00000513179.1
ENST00000515205.1 |
RP11-539L10.3
|
RP11-539L10.3 |
| chr9_+_69650263 | 0.58 |
ENST00000322495.3
|
AL445665.1
|
Protein LOC100996643 |
| chr4_+_165675269 | 0.55 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr8_-_110988070 | 0.52 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr3_+_181429704 | 0.48 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr15_+_76352178 | 0.47 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr13_+_96743093 | 0.46 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr14_-_81902516 | 0.42 |
ENST00000554710.1
|
STON2
|
stonin 2 |
| chr8_-_33330595 | 0.41 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr22_-_22090043 | 0.41 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr16_-_30022293 | 0.40 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr5_-_73936544 | 0.40 |
ENST00000509127.2
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chrX_+_55744228 | 0.38 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr1_-_219615984 | 0.37 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr10_+_35484793 | 0.35 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr14_-_81902791 | 0.35 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr17_-_39258461 | 0.34 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr13_+_20268547 | 0.34 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr20_-_50159198 | 0.32 |
ENST00000371564.3
ENST00000396009.3 ENST00000610033.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr1_+_156308245 | 0.32 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr1_+_33283043 | 0.32 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr1_-_95392635 | 0.30 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr1_+_222791417 | 0.29 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr5_+_15500280 | 0.28 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr6_-_27440460 | 0.28 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_-_33283754 | 0.28 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr16_-_25122785 | 0.27 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr15_-_76352069 | 0.27 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr17_+_19920456 | 0.26 |
ENST00000582604.1
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr6_-_27440837 | 0.26 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr12_+_96337061 | 0.25 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr17_+_78194205 | 0.25 |
ENST00000573809.1
ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr5_-_73936451 | 0.25 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr4_-_90757364 | 0.25 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_+_98881268 | 0.24 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr5_-_59064458 | 0.24 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_+_12726623 | 0.23 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr11_+_44748361 | 0.23 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr15_-_30114231 | 0.22 |
ENST00000356107.6
ENST00000545208.2 |
TJP1
|
tight junction protein 1 |
| chr3_+_32280159 | 0.22 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr15_-_98646940 | 0.22 |
ENST00000560195.1
|
CTD-2544M6.2
|
CTD-2544M6.2 |
| chr16_-_103572 | 0.22 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr1_+_109289279 | 0.22 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr7_+_154795154 | 0.22 |
ENST00000608317.1
|
PAXIP1-AS1
|
PAXIP1 antisense RNA 1 (head to head) |
| chr3_-_139195350 | 0.21 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr1_-_104239302 | 0.21 |
ENST00000446703.1
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr1_+_104197912 | 0.21 |
ENST00000430659.1
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr9_+_125703282 | 0.21 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr19_-_5838734 | 0.20 |
ENST00000532464.1
ENST00000528505.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr1_+_16083123 | 0.20 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_-_33049690 | 0.19 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr16_+_67063855 | 0.19 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr2_-_24583314 | 0.19 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr4_-_120133661 | 0.18 |
ENST00000503243.1
ENST00000326780.3 |
RP11-455G16.1
|
Uncharacterized protein |
| chr2_-_224467093 | 0.18 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr15_+_63889577 | 0.17 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr17_-_34257771 | 0.17 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr1_-_21044489 | 0.17 |
ENST00000247986.2
|
KIF17
|
kinesin family member 17 |
| chr1_-_9129598 | 0.17 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr3_-_42846021 | 0.17 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr22_+_40742512 | 0.17 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr1_+_55271736 | 0.17 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr11_-_85430356 | 0.17 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_41258786 | 0.17 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_-_42845951 | 0.17 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr1_-_247335269 | 0.17 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr3_+_151986709 | 0.16 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr7_+_130126165 | 0.16 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr1_-_104239076 | 0.16 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr15_-_30113676 | 0.16 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr1_+_156308403 | 0.16 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr18_+_47087390 | 0.16 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr9_+_214842 | 0.16 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr15_+_63889552 | 0.16 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chrX_-_134232630 | 0.16 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr10_+_6779326 | 0.16 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr8_-_101718991 | 0.16 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr10_-_95209 | 0.15 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr6_+_90272027 | 0.15 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr6_-_31080336 | 0.15 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr8_+_82644669 | 0.15 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr15_-_34628951 | 0.15 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_+_173763250 | 0.15 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr1_+_16083098 | 0.15 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr14_-_77495007 | 0.14 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr15_-_44486632 | 0.14 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr22_+_40742497 | 0.14 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr1_+_222988464 | 0.14 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr22_-_22090064 | 0.14 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chrX_-_31284974 | 0.14 |
ENST00000378702.4
|
DMD
|
dystrophin |
| chr14_+_23776167 | 0.14 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr5_+_74011328 | 0.14 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr1_-_9129631 | 0.14 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr21_-_35987438 | 0.13 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr19_-_49250054 | 0.13 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr6_+_121756809 | 0.12 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr19_-_12624647 | 0.12 |
ENST00000455490.1
|
ZNF709
|
zinc finger protein 709 |
| chr1_-_151813033 | 0.12 |
ENST00000454109.1
|
C2CD4D
|
C2 calcium-dependent domain containing 4D |
| chr1_+_33283246 | 0.12 |
ENST00000526230.1
ENST00000531256.1 ENST00000482212.1 |
S100PBP
|
S100P binding protein |
| chr4_-_89619386 | 0.12 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr21_-_34852304 | 0.12 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr16_-_53537105 | 0.12 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr16_+_2198604 | 0.12 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr9_+_74526532 | 0.11 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr1_-_104238912 | 0.11 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr12_-_15374328 | 0.11 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr7_+_130126012 | 0.10 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chrX_-_31285018 | 0.10 |
ENST00000361471.4
|
DMD
|
dystrophin |
| chr11_+_27062272 | 0.10 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr2_+_149402009 | 0.10 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr5_+_32710736 | 0.10 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr16_-_50402690 | 0.10 |
ENST00000394689.2
|
BRD7
|
bromodomain containing 7 |
| chr7_+_12727250 | 0.10 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr12_-_122907091 | 0.10 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr5_+_138611798 | 0.10 |
ENST00000502394.1
|
MATR3
|
matrin 3 |
| chr10_+_99894399 | 0.10 |
ENST00000298999.3
ENST00000314594.5 |
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr13_+_39261224 | 0.10 |
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2 |
| chr4_-_108204846 | 0.09 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr4_-_149363662 | 0.09 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr4_-_52904425 | 0.09 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr19_+_30097181 | 0.09 |
ENST00000586420.1
ENST00000221770.3 ENST00000392279.3 ENST00000590688.1 |
POP4
|
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr5_+_138089100 | 0.09 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr5_-_151066514 | 0.09 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr7_-_122526499 | 0.09 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr17_-_1029866 | 0.09 |
ENST00000570525.1
|
ABR
|
active BCR-related |
| chr6_+_7107999 | 0.09 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_44115814 | 0.09 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr11_+_32112431 | 0.09 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chrX_-_31285042 | 0.09 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr20_-_45061695 | 0.09 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chr17_-_33760269 | 0.09 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr12_+_16500037 | 0.08 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr20_-_23402028 | 0.08 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr4_-_83483395 | 0.08 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr7_+_138145145 | 0.08 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr2_-_96192450 | 0.08 |
ENST00000609975.1
|
RP11-440D17.3
|
RP11-440D17.3 |
| chr16_-_82045049 | 0.08 |
ENST00000532128.1
ENST00000328945.5 |
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr2_-_180726232 | 0.08 |
ENST00000410066.1
|
ZNF385B
|
zinc finger protein 385B |
| chr12_+_50794947 | 0.08 |
ENST00000552445.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr19_-_5838768 | 0.08 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr1_-_9129895 | 0.08 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr4_-_149363376 | 0.08 |
ENST00000512865.1
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr17_-_33760164 | 0.08 |
ENST00000445092.1
ENST00000394562.1 ENST00000447040.2 |
SLFN12
|
schlafen family member 12 |
| chr1_+_67395922 | 0.08 |
ENST00000401042.3
ENST00000355356.3 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr2_+_223162866 | 0.07 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr2_-_175870085 | 0.07 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr11_-_6624801 | 0.07 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr10_+_99894380 | 0.07 |
ENST00000370584.3
|
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr17_+_67410832 | 0.07 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr6_+_28092338 | 0.07 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr9_+_99212403 | 0.07 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr17_-_10101868 | 0.07 |
ENST00000432992.2
ENST00000540214.1 |
GAS7
|
growth arrest-specific 7 |
| chr17_-_44657017 | 0.07 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr5_-_111091948 | 0.06 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr4_-_170897045 | 0.06 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr9_+_74526384 | 0.06 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr5_+_125758865 | 0.06 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr14_-_64761078 | 0.06 |
ENST00000341099.4
ENST00000556275.1 ENST00000542956.1 ENST00000353772.3 ENST00000357782.2 ENST00000267525.6 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr9_+_100174344 | 0.06 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr11_+_125461607 | 0.06 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr6_-_135818368 | 0.06 |
ENST00000367798.2
|
AHI1
|
Abelson helper integration site 1 |
| chr12_-_71148357 | 0.06 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_54684327 | 0.06 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_+_97515409 | 0.06 |
ENST00000371207.3
ENST00000543964.1 |
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr4_-_5890145 | 0.06 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr3_-_62860704 | 0.06 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr10_+_90672113 | 0.06 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr7_+_94536898 | 0.06 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr6_-_42418999 | 0.06 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr1_+_24645915 | 0.06 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_+_125758813 | 0.06 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr19_-_42759300 | 0.06 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr14_-_102829051 | 0.06 |
ENST00000536961.2
ENST00000541568.2 ENST00000216756.6 |
CINP
|
cyclin-dependent kinase 2 interacting protein |
| chr2_+_62423242 | 0.06 |
ENST00000301998.4
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chrX_-_153141783 | 0.06 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr16_+_25123148 | 0.06 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr12_-_67197760 | 0.06 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr10_+_24498060 | 0.06 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr1_+_222988406 | 0.06 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr17_+_63133587 | 0.05 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr2_-_241759622 | 0.05 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr17_+_53343577 | 0.05 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr12_-_31478428 | 0.05 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr16_+_25123041 | 0.05 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr1_+_104198377 | 0.05 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr12_-_56221330 | 0.05 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr17_+_63133526 | 0.05 |
ENST00000443584.3
|
RGS9
|
regulator of G-protein signaling 9 |
| chr17_-_32484313 | 0.05 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr1_-_154928562 | 0.05 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr14_-_71107921 | 0.05 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr3_+_167453026 | 0.05 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_-_167232484 | 0.05 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr8_-_11873043 | 0.05 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr10_+_24497704 | 0.05 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chrX_-_153141434 | 0.05 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr17_-_34257731 | 0.05 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr17_-_40288449 | 0.05 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr5_-_114631958 | 0.05 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr4_-_110223523 | 0.05 |
ENST00000399127.1
|
COL25A1
|
collagen, type XXV, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.7 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0039020 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric nephron tubule development(GO:0039020) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.0 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.6 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |