Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
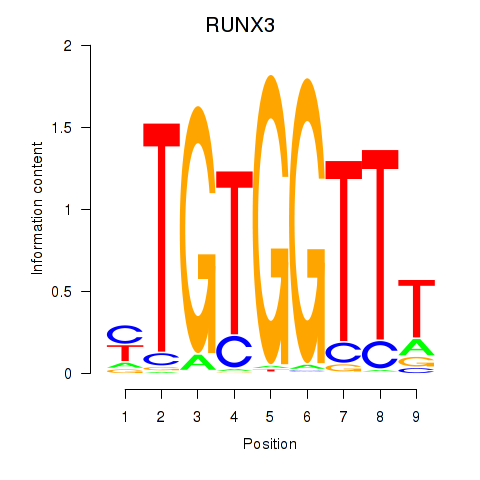
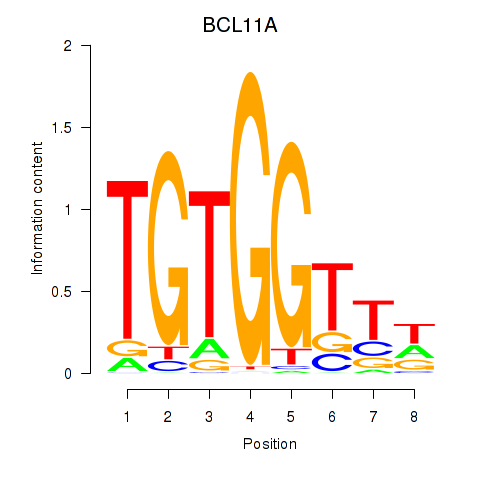
Results for RUNX3_BCL11A
Z-value: 1.13
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL11A | hg19_v2_chr2_-_60780536_60780541 | -0.77 | 7.1e-02 | Click! |
| RUNX3 | hg19_v2_chr1_-_25291475_25291511 | 0.42 | 4.0e-01 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_131409476 | 3.04 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr5_+_159895275 | 1.08 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr5_+_35856951 | 0.95 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr1_-_235098861 | 0.91 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr2_+_228678550 | 0.84 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr15_+_67390920 | 0.79 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr6_+_45296048 | 0.64 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr20_+_44637526 | 0.61 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_+_45296391 | 0.56 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr17_+_76037081 | 0.56 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr2_-_214017151 | 0.55 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chrX_-_19688475 | 0.53 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr6_+_30029008 | 0.49 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr19_-_42636617 | 0.48 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr19_-_8567505 | 0.47 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr21_-_36421401 | 0.47 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr15_-_60695071 | 0.46 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr2_+_33359473 | 0.45 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr14_-_74462922 | 0.44 |
ENST00000553284.1
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr15_+_86098670 | 0.42 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr22_-_30642782 | 0.41 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr1_+_152486950 | 0.41 |
ENST00000368790.3
|
CRCT1
|
cysteine-rich C-terminal 1 |
| chr12_-_96793142 | 0.37 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr22_-_30642728 | 0.37 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr12_-_58145889 | 0.37 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr6_+_31543334 | 0.34 |
ENST00000449264.2
|
TNF
|
tumor necrosis factor |
| chr9_-_117853297 | 0.34 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr11_-_58345569 | 0.34 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr1_+_156308245 | 0.33 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr2_-_61389168 | 0.33 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr1_+_164528437 | 0.32 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr12_-_4754339 | 0.32 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr16_+_82090028 | 0.30 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_-_25291475 | 0.29 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr1_+_110453608 | 0.29 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr19_-_42636543 | 0.29 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr1_-_235098935 | 0.28 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr2_+_173955327 | 0.28 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr7_+_18535321 | 0.28 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr17_+_7239821 | 0.28 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr3_+_50126341 | 0.28 |
ENST00000347869.3
ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5
|
RNA binding motif protein 5 |
| chr9_+_75766763 | 0.28 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr17_+_65375082 | 0.27 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr5_-_56778635 | 0.26 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr9_+_75766652 | 0.25 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chr14_+_75988851 | 0.25 |
ENST00000555504.1
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr14_+_66578299 | 0.25 |
ENST00000554187.1
ENST00000556662.1 ENST00000556291.1 ENST00000557723.1 ENST00000557050.1 |
RP11-783L4.1
|
RP11-783L4.1 |
| chr17_-_69198295 | 0.24 |
ENST00000569074.1
|
CASC17
|
cancer susceptibility candidate 17 (non-protein coding) |
| chr17_+_27047570 | 0.24 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chr1_+_209859510 | 0.23 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr1_-_92951607 | 0.23 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr5_+_67535647 | 0.23 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr20_+_36405665 | 0.22 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr1_+_53098862 | 0.22 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr3_+_49977440 | 0.22 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr10_+_17272608 | 0.22 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr3_+_98699880 | 0.21 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chrX_-_54209640 | 0.21 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr3_+_51575596 | 0.21 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr21_-_31588365 | 0.21 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr11_-_128457446 | 0.21 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr5_+_148206156 | 0.20 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr7_+_102389434 | 0.20 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr3_-_99569821 | 0.20 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_-_131756559 | 0.20 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr17_-_61777090 | 0.19 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr6_+_12290586 | 0.19 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr4_+_76481258 | 0.19 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr6_-_152623231 | 0.19 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_153348067 | 0.18 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr2_-_61389240 | 0.18 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr6_-_31508304 | 0.18 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr17_+_7239904 | 0.18 |
ENST00000575425.1
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr16_-_69418553 | 0.18 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr6_-_75912508 | 0.18 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr11_-_102826434 | 0.18 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr3_-_100558953 | 0.17 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr11_-_71781096 | 0.17 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_18741882 | 0.17 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr22_+_31488433 | 0.17 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr16_+_84801852 | 0.17 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr17_-_73840614 | 0.17 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_+_19281787 | 0.17 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_+_139135648 | 0.17 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr3_+_15643140 | 0.17 |
ENST00000449107.1
|
BTD
|
biotinidase |
| chr15_-_72668185 | 0.16 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr1_+_111772314 | 0.16 |
ENST00000466741.1
ENST00000477185.2 |
CHI3L2
|
chitinase 3-like 2 |
| chr14_-_23564320 | 0.16 |
ENST00000605057.1
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr1_-_161039647 | 0.16 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr3_+_49977523 | 0.16 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr1_-_161039753 | 0.16 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_-_217250231 | 0.16 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr13_-_99910673 | 0.16 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr6_-_112081113 | 0.16 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr6_+_15401075 | 0.16 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr8_-_60031762 | 0.15 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr1_+_111772375 | 0.15 |
ENST00000467038.2
ENST00000497587.2 |
CHI3L2
|
chitinase 3-like 2 |
| chr2_+_33359687 | 0.15 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr5_-_127674883 | 0.15 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr8_-_72274095 | 0.15 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr2_+_33359646 | 0.15 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_155099927 | 0.15 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr2_+_234545148 | 0.15 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_73461364 | 0.15 |
ENST00000540468.1
ENST00000539919.1 ENST00000258091.5 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr21_-_31588338 | 0.14 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr3_+_49977490 | 0.14 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr1_+_149553003 | 0.14 |
ENST00000369222.3
|
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4C |
| chr3_+_40518599 | 0.14 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr17_-_61776522 | 0.14 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr1_-_144364246 | 0.14 |
ENST00000540273.1
|
PPIAL4B
|
peptidylprolyl isomerase A (cyclophilin A)-like 4B |
| chr16_+_58033450 | 0.14 |
ENST00000561743.1
|
USB1
|
U6 snRNA biogenesis 1 |
| chr3_+_14061155 | 0.14 |
ENST00000532924.1
|
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr10_+_25463951 | 0.13 |
ENST00000376351.3
|
GPR158
|
G protein-coupled receptor 158 |
| chr1_+_155579979 | 0.13 |
ENST00000452804.2
ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr17_-_7164410 | 0.13 |
ENST00000574070.1
|
CLDN7
|
claudin 7 |
| chr1_-_161039456 | 0.13 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr13_-_46756351 | 0.13 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_110453109 | 0.13 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr2_+_169659121 | 0.13 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr7_-_10979750 | 0.13 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr2_+_73461410 | 0.13 |
ENST00000399032.2
ENST00000398422.2 ENST00000537131.1 ENST00000538797.1 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr12_-_16760021 | 0.13 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr14_-_21493884 | 0.13 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr5_+_145316120 | 0.13 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr7_-_87104963 | 0.13 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr12_-_4754318 | 0.13 |
ENST00000536414.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr9_-_130541017 | 0.13 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr15_+_67418047 | 0.13 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_+_18330035 | 0.12 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr4_+_89299885 | 0.12 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr14_+_35761580 | 0.12 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr2_-_71221942 | 0.12 |
ENST00000272438.4
|
TEX261
|
testis expressed 261 |
| chr17_+_9745786 | 0.12 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr14_-_21493649 | 0.12 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr15_-_78358465 | 0.12 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr9_-_123342415 | 0.12 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr17_+_7462103 | 0.12 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_-_106146547 | 0.12 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr1_+_209929377 | 0.12 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr16_+_67312049 | 0.12 |
ENST00000565899.1
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr7_-_99679324 | 0.12 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr3_-_49941042 | 0.12 |
ENST00000344206.4
ENST00000296474.3 |
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr18_-_43678241 | 0.12 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr2_+_44001172 | 0.11 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr2_+_169658928 | 0.11 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_+_142342240 | 0.11 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr9_-_93925369 | 0.11 |
ENST00000457976.1
|
RP11-305L7.3
|
RP11-305L7.3 |
| chr18_+_5238549 | 0.11 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr1_+_110453462 | 0.11 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr10_-_99205607 | 0.11 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr17_+_38673270 | 0.11 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr11_+_71238313 | 0.11 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr6_+_30294612 | 0.11 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr6_+_37787704 | 0.11 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr3_+_15643245 | 0.11 |
ENST00000303498.5
ENST00000437172.1 |
BTD
|
biotinidase |
| chr11_+_60739115 | 0.11 |
ENST00000344028.5
ENST00000346437.4 |
CD6
|
CD6 molecule |
| chr10_+_89420706 | 0.11 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_+_30455016 | 0.11 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chr6_+_27925019 | 0.11 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr6_+_30295036 | 0.11 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr20_-_25062767 | 0.11 |
ENST00000429762.3
ENST00000444511.2 ENST00000376707.3 |
VSX1
|
visual system homeobox 1 |
| chr4_+_89299994 | 0.10 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_-_50868882 | 0.10 |
ENST00000598915.1
|
NAPSA
|
napsin A aspartic peptidase |
| chr19_-_49496557 | 0.10 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr12_-_49245916 | 0.10 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr11_+_61015594 | 0.10 |
ENST00000451616.2
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr17_-_42188598 | 0.10 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr15_-_37393406 | 0.10 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr16_+_72088376 | 0.10 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr15_+_63889552 | 0.10 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr3_-_71179699 | 0.10 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr12_-_49245936 | 0.10 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr17_-_56494713 | 0.10 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr9_-_34048873 | 0.10 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr3_+_15643476 | 0.10 |
ENST00000436193.1
ENST00000383778.4 |
BTD
|
biotinidase |
| chrX_-_119005735 | 0.10 |
ENST00000371442.2
|
RNF113A
|
ring finger protein 113A |
| chrX_+_37639302 | 0.10 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr15_-_41805994 | 0.09 |
ENST00000561619.1
ENST00000263800.6 ENST00000355166.5 ENST00000453182.2 |
LTK
|
leukocyte receptor tyrosine kinase |
| chr4_+_152020736 | 0.09 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr3_+_187957646 | 0.09 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr1_-_86043921 | 0.09 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr5_+_132149017 | 0.09 |
ENST00000378693.2
|
SOWAHA
|
sosondowah ankyrin repeat domain family member A |
| chr9_-_130517522 | 0.09 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr12_-_16759440 | 0.09 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_+_5030205 | 0.09 |
ENST00000606761.1
ENST00000607327.1 ENST00000606767.1 |
RP11-428J1.5
|
RP11-428J1.5 |
| chr11_+_62475130 | 0.09 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr12_+_57916466 | 0.09 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr2_-_74779744 | 0.09 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr6_-_34524049 | 0.09 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr3_+_42892458 | 0.09 |
ENST00000494619.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr1_-_94147385 | 0.09 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_-_51717875 | 0.09 |
ENST00000604560.1
|
BIN2
|
bridging integrator 2 |
| chr12_-_51717948 | 0.09 |
ENST00000267012.4
|
BIN2
|
bridging integrator 2 |
| chr15_+_63889577 | 0.09 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr1_+_207238326 | 0.09 |
ENST00000541914.1
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr12_-_51717922 | 0.09 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr5_-_65018834 | 0.09 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_+_31497271 | 0.09 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr19_-_40786733 | 0.09 |
ENST00000486368.2
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_+_172628154 | 0.08 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 0.8 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.5 | GO:0031394 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.5 | GO:0052361 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.6 | GO:1904141 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 1.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.3 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0060585 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.6 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.1 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) |
| 0.0 | 0.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.4 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 0.1 | GO:2000525 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 1.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.2 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.3 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0002884 | negative regulation of type IV hypersensitivity(GO:0001808) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:1990791 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.0 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 0.8 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 0.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 2.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |