Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
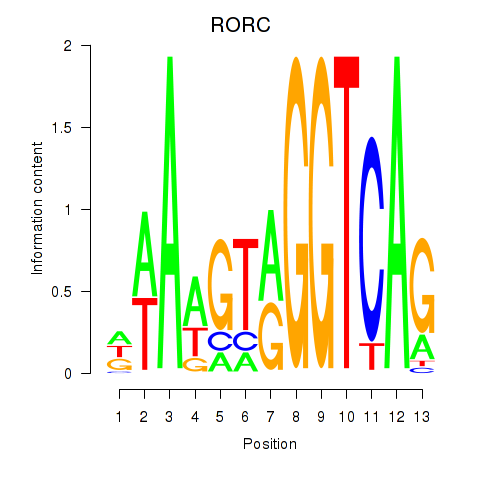
Results for RORC
Z-value: 0.35
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORC | hg19_v2_chr1_-_151804314_151804348 | -0.41 | 4.2e-01 | Click! |
Activity profile of RORC motif
Sorted Z-values of RORC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_71485681 | 0.34 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr14_-_68157084 | 0.22 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr19_+_39759154 | 0.18 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chrX_+_69501943 | 0.17 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr8_-_95449155 | 0.16 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr9_-_35042824 | 0.16 |
ENST00000595331.1
|
FLJ00273
|
FLJ00273 |
| chr17_+_47439733 | 0.16 |
ENST00000507337.1
|
RP11-1079K10.3
|
RP11-1079K10.3 |
| chr17_-_46623441 | 0.15 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr4_+_85504075 | 0.15 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr19_-_49140609 | 0.14 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr1_-_211752073 | 0.14 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chrX_-_102531717 | 0.14 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr2_+_160590469 | 0.12 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr20_-_44007014 | 0.12 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr14_-_54418598 | 0.11 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr19_-_49140692 | 0.10 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr3_-_45957088 | 0.10 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr19_-_46580352 | 0.10 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr14_+_23025534 | 0.09 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr8_+_124780672 | 0.08 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr11_+_13299186 | 0.08 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr12_+_4671352 | 0.08 |
ENST00000542744.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_-_45957534 | 0.08 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_-_148778323 | 0.08 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr6_+_89855765 | 0.08 |
ENST00000275072.4
|
PM20D2
|
peptidase M20 domain containing 2 |
| chr3_+_187461442 | 0.08 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr11_-_10822029 | 0.07 |
ENST00000528839.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_+_44679808 | 0.07 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr18_-_23671139 | 0.07 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr4_-_1188922 | 0.06 |
ENST00000515004.1
ENST00000502483.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr16_-_69385681 | 0.05 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr17_-_37764128 | 0.05 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr1_-_157069590 | 0.05 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr4_-_87770416 | 0.05 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr7_+_112063192 | 0.04 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr14_-_23451467 | 0.04 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr19_-_10450287 | 0.04 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr12_+_93963590 | 0.03 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr3_+_160559931 | 0.03 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr1_-_150207017 | 0.03 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_140586922 | 0.03 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr13_+_109248500 | 0.03 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr2_+_162016827 | 0.03 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_159094194 | 0.03 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr10_-_103578182 | 0.03 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr2_+_162016916 | 0.03 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_217311090 | 0.02 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr1_+_171810606 | 0.02 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr14_-_23755297 | 0.02 |
ENST00000357460.5
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr17_-_48785216 | 0.02 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr1_+_114447763 | 0.02 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr10_-_105845674 | 0.02 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr12_+_56414795 | 0.02 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr1_-_62190793 | 0.02 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr1_+_205197304 | 0.01 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr1_+_104198377 | 0.01 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr4_-_170948361 | 0.01 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_153522562 | 0.01 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr17_-_10707410 | 0.01 |
ENST00000581851.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
| chr1_+_154540246 | 0.01 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr10_-_103578162 | 0.01 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr1_-_145826450 | 0.01 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr9_-_110251836 | 0.01 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr1_+_26869597 | 0.01 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr6_-_111927062 | 0.01 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr1_-_185286461 | 0.00 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr12_+_93964158 | 0.00 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr3_-_65583561 | 0.00 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr14_-_23451845 | 0.00 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:1905072 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |