Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
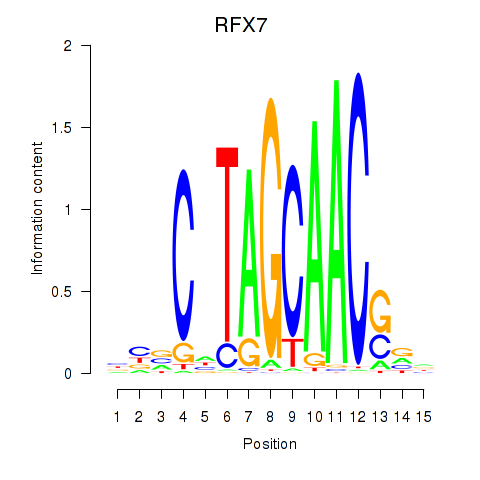
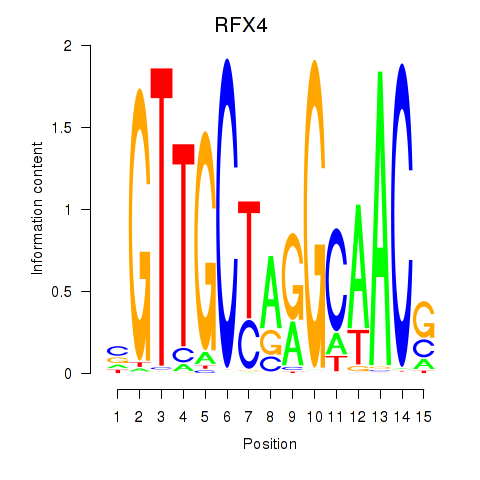
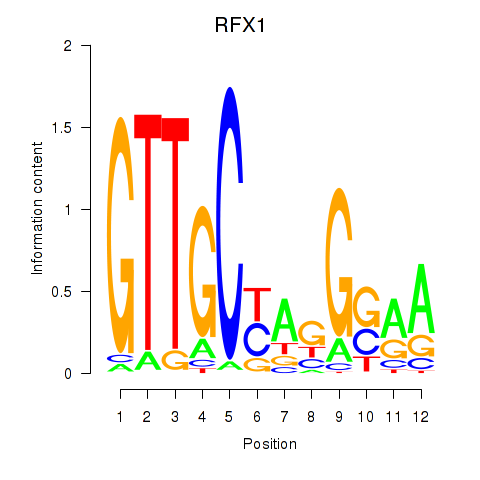
Results for RFX7_RFX4_RFX1
Z-value: 1.33
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | 0.75 | 8.6e-02 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | 0.57 | 2.3e-01 | Click! |
| RFX4 | hg19_v2_chr12_+_106994905_106994954 | -0.09 | 8.6e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51014460 | 2.56 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_-_51014588 | 1.64 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_+_55996565 | 1.07 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr19_-_51014345 | 0.97 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr16_+_24550857 | 0.76 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr19_-_50316517 | 0.74 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr19_-_50316489 | 0.71 |
ENST00000533418.1
|
FUZ
|
fuzzy planar cell polarity protein |
| chr7_-_105221898 | 0.69 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr13_+_103046954 | 0.67 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr1_+_161068179 | 0.66 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr19_+_507299 | 0.63 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr1_-_85155939 | 0.55 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr19_-_50316423 | 0.54 |
ENST00000528094.1
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr1_+_32674675 | 0.54 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr12_+_119772502 | 0.52 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr12_+_119772734 | 0.49 |
ENST00000539847.1
|
CCDC60
|
coiled-coil domain containing 60 |
| chr1_-_86861660 | 0.45 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr19_-_40324767 | 0.44 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_-_55791058 | 0.42 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr4_-_2420335 | 0.41 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr22_-_23484246 | 0.41 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr6_+_89790490 | 0.40 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr17_-_15244894 | 0.40 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr4_-_156297949 | 0.39 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr11_+_61248583 | 0.39 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr17_+_48585794 | 0.37 |
ENST00000576179.1
ENST00000419930.1 |
MYCBPAP
|
MYCBP associated protein |
| chr1_-_74663825 | 0.36 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chrX_+_23925918 | 0.36 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr12_-_121973974 | 0.36 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_-_156298028 | 0.36 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr19_-_41220957 | 0.34 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr1_+_74663994 | 0.33 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr1_-_85156216 | 0.33 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr19_+_55996316 | 0.33 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr9_+_26956371 | 0.33 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr7_+_12726474 | 0.33 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr1_+_36549676 | 0.32 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr10_+_106113515 | 0.32 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr14_+_89290965 | 0.32 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr19_+_48972459 | 0.31 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr3_+_158449972 | 0.31 |
ENST00000486568.1
|
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr2_-_27851745 | 0.31 |
ENST00000394775.3
ENST00000522876.1 |
CCDC121
|
coiled-coil domain containing 121 |
| chr16_-_29875057 | 0.31 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr20_-_44420507 | 0.31 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr10_-_25304889 | 0.31 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr14_+_52313833 | 0.31 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_85156090 | 0.31 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_+_43637996 | 0.30 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr7_+_23637118 | 0.30 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr10_+_15001430 | 0.30 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr16_+_30773610 | 0.30 |
ENST00000566811.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr19_-_40324255 | 0.29 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr9_-_117267717 | 0.29 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr4_-_156298087 | 0.28 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr1_+_6640046 | 0.28 |
ENST00000319084.5
ENST00000435905.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr17_+_42977122 | 0.28 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr12_-_76478386 | 0.27 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_+_5914213 | 0.27 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr6_+_89790459 | 0.27 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr14_+_105452094 | 0.26 |
ENST00000551606.1
ENST00000547315.1 |
C14orf79
|
chromosome 14 open reading frame 79 |
| chr18_-_32957260 | 0.26 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chr10_+_70980051 | 0.26 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr5_-_54529415 | 0.26 |
ENST00000282572.4
|
CCNO
|
cyclin O |
| chr3_-_48659193 | 0.26 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr11_+_450255 | 0.26 |
ENST00000308020.5
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr2_+_172778952 | 0.26 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr17_+_29815113 | 0.26 |
ENST00000583755.1
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr1_-_45140227 | 0.25 |
ENST00000372237.3
|
TMEM53
|
transmembrane protein 53 |
| chr4_-_140005341 | 0.25 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr11_+_537494 | 0.24 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr20_+_17550489 | 0.24 |
ENST00000246069.7
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr17_+_2264983 | 0.24 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chrX_-_131623874 | 0.24 |
ENST00000436215.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr11_-_73882249 | 0.23 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr5_-_149669612 | 0.23 |
ENST00000510347.1
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr2_-_63815628 | 0.23 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr16_+_74411776 | 0.22 |
ENST00000429990.1
|
NPIPB15
|
nuclear pore complex interacting protein family, member B15 |
| chr16_-_72127550 | 0.22 |
ENST00000268483.3
|
TXNL4B
|
thioredoxin-like 4B |
| chr14_+_96343100 | 0.22 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr3_-_195076933 | 0.22 |
ENST00000423531.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr18_+_29671812 | 0.21 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr17_-_65242063 | 0.21 |
ENST00000581159.1
|
HELZ
|
helicase with zinc finger |
| chr11_-_559377 | 0.20 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr2_-_196933536 | 0.20 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr11_-_108422926 | 0.20 |
ENST00000428840.1
ENST00000526312.1 |
EXPH5
|
exophilin 5 |
| chr20_+_306177 | 0.20 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr19_-_11545920 | 0.19 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr1_-_63988846 | 0.19 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr16_-_71598823 | 0.19 |
ENST00000566202.1
|
ZNF19
|
zinc finger protein 19 |
| chr3_+_132379154 | 0.19 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr11_+_65779283 | 0.19 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr1_+_63989004 | 0.19 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr16_+_50059182 | 0.19 |
ENST00000562576.1
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr5_-_138739739 | 0.19 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr8_+_86999516 | 0.18 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr8_+_6566206 | 0.18 |
ENST00000518327.1
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr5_-_133702761 | 0.18 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr1_-_51791596 | 0.18 |
ENST00000532836.1
ENST00000422925.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr12_-_76478417 | 0.18 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_+_111051832 | 0.18 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr17_+_48585958 | 0.18 |
ENST00000436259.2
|
MYCBPAP
|
MYCBP associated protein |
| chr11_+_71791693 | 0.18 |
ENST00000289488.2
ENST00000447974.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr5_+_110074685 | 0.18 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr4_-_140544386 | 0.18 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr12_-_76478446 | 0.18 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr8_+_15397732 | 0.18 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr11_-_66104237 | 0.18 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr20_-_49575081 | 0.17 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr2_-_62081148 | 0.17 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr4_+_926171 | 0.17 |
ENST00000507319.1
ENST00000264771.4 |
TMEM175
|
transmembrane protein 175 |
| chr9_-_130477912 | 0.17 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr14_-_22005062 | 0.17 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_40627038 | 0.17 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr20_-_49575058 | 0.17 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr11_-_64885111 | 0.17 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr1_-_36915880 | 0.17 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr4_-_153457197 | 0.16 |
ENST00000281708.4
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr19_+_58111241 | 0.16 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr13_+_21141270 | 0.16 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr17_-_80656528 | 0.16 |
ENST00000538809.2
ENST00000269347.6 ENST00000571995.1 |
RAB40B
|
RAB40B, member RAS oncogene family |
| chr15_+_49913175 | 0.16 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr1_+_24120143 | 0.16 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr20_+_17550691 | 0.16 |
ENST00000474024.1
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr3_-_10149907 | 0.16 |
ENST00000450660.2
ENST00000524279.1 |
FANCD2OS
|
FANCD2 opposite strand |
| chr6_+_112408768 | 0.16 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr2_-_62081254 | 0.16 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr19_+_41856816 | 0.16 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr20_+_18447771 | 0.16 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr15_+_49913201 | 0.16 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr2_+_95537248 | 0.16 |
ENST00000427593.2
|
TEKT4
|
tektin 4 |
| chr15_+_40531621 | 0.16 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr12_-_76478686 | 0.15 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_+_11546153 | 0.15 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr15_+_40531243 | 0.15 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr21_-_43916296 | 0.15 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr2_-_192016276 | 0.15 |
ENST00000413064.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr6_+_134274322 | 0.15 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr19_+_50321528 | 0.15 |
ENST00000312865.6
ENST00000595185.1 ENST00000538643.1 |
MED25
|
mediator complex subunit 25 |
| chr4_-_177116772 | 0.15 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr19_-_55791431 | 0.15 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_-_1051455 | 0.15 |
ENST00000379339.1
ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr11_+_86748957 | 0.15 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chrX_-_114252193 | 0.15 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr11_-_66103932 | 0.14 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_-_74648702 | 0.14 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr21_-_47743719 | 0.14 |
ENST00000397680.1
ENST00000445935.1 ENST00000397685.4 ENST00000397682.3 ENST00000291691.7 |
C21orf58
|
chromosome 21 open reading frame 58 |
| chr7_+_156902674 | 0.14 |
ENST00000594086.1
|
AC006967.1
|
Protein LOC100996426 |
| chr8_-_144623595 | 0.14 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr12_-_85430024 | 0.14 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr20_+_814349 | 0.14 |
ENST00000381941.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr19_+_14184370 | 0.14 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr19_-_51017881 | 0.14 |
ENST00000601207.1
ENST00000598657.1 ENST00000376916.3 |
ASPDH
|
aspartate dehydrogenase domain containing |
| chr20_+_306221 | 0.14 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr17_+_48585745 | 0.14 |
ENST00000323776.5
|
MYCBPAP
|
MYCBP associated protein |
| chr6_+_116937636 | 0.14 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr15_-_49447771 | 0.14 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr19_-_5293243 | 0.13 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr1_+_85527987 | 0.13 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr6_-_46620522 | 0.13 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr10_-_82116497 | 0.13 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr18_+_61254221 | 0.13 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr17_+_78010428 | 0.13 |
ENST00000397545.4
ENST00000374877.3 ENST00000269318.5 ENST00000374876.4 ENST00000576033.1 ENST00000574099.1 |
CCDC40
|
coiled-coil domain containing 40 |
| chr15_+_71184931 | 0.13 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr22_-_33968239 | 0.13 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr19_+_5681153 | 0.13 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr15_+_81489213 | 0.13 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr6_-_80247140 | 0.13 |
ENST00000392959.1
ENST00000467898.3 |
LCA5
|
Leber congenital amaurosis 5 |
| chr3_-_149510553 | 0.13 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr19_+_55888186 | 0.13 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr1_+_111888890 | 0.13 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr22_-_39268192 | 0.13 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr3_+_149689066 | 0.13 |
ENST00000593416.1
|
AC117395.1
|
LOC646903 protein; Uncharacterized protein |
| chr12_+_49297887 | 0.13 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr3_-_47324079 | 0.13 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr3_-_45883558 | 0.13 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr12_+_14927270 | 0.13 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr5_+_176873789 | 0.12 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr1_-_118727781 | 0.12 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr1_-_1051736 | 0.12 |
ENST00000448924.1
ENST00000294576.5 ENST00000437760.1 ENST00000462097.1 ENST00000475119.1 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr12_+_111051902 | 0.12 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr8_+_38065104 | 0.12 |
ENST00000521311.1
|
BAG4
|
BCL2-associated athanogene 4 |
| chr14_-_74025625 | 0.12 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr19_-_59084647 | 0.12 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr12_-_108154705 | 0.12 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr16_-_88923285 | 0.12 |
ENST00000542788.1
ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chrX_-_131623982 | 0.12 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr6_+_52285131 | 0.12 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr15_+_85923856 | 0.12 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_+_74663896 | 0.12 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr16_+_30772913 | 0.12 |
ENST00000563909.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr4_+_26585538 | 0.12 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr19_+_35634146 | 0.11 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr19_+_11546440 | 0.11 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr7_+_2598628 | 0.11 |
ENST00000402050.2
ENST00000404984.1 ENST00000415271.2 ENST00000438376.2 |
IQCE
|
IQ motif containing E |
| chr19_+_16308711 | 0.11 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr16_+_78133536 | 0.11 |
ENST00000402655.2
ENST00000406884.2 ENST00000539474.2 ENST00000569818.1 ENST00000355860.3 ENST00000408984.3 |
WWOX
|
WW domain containing oxidoreductase |
| chr2_+_26785409 | 0.11 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr1_-_207226313 | 0.11 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr14_+_61447832 | 0.11 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr15_+_71185148 | 0.11 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.4 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.1 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.2 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.6 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0031329 | regulation of cellular catabolic process(GO:0031329) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.2 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.0 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.0 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.0 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 5.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 1.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |