Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
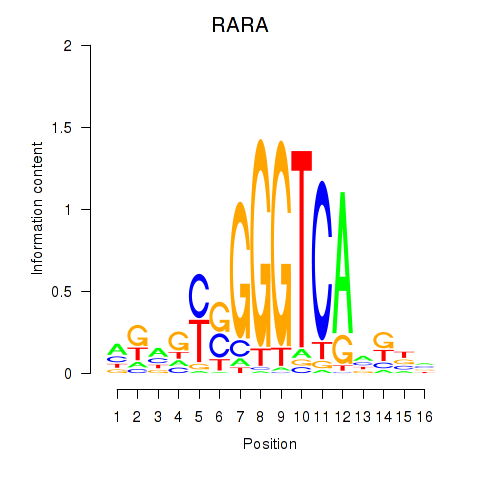
Results for RARA
Z-value: 0.72
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | retinoic acid receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38498594_38498661 | -0.64 | 1.7e-01 | Click! |
Activity profile of RARA motif
Sorted Z-values of RARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_70963538 | 0.59 |
ENST00000413503.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_-_72374848 | 0.58 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr6_+_151358048 | 0.51 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr14_+_38065052 | 0.43 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chrX_-_37706815 | 0.42 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr14_-_54955376 | 0.39 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr5_-_136649218 | 0.39 |
ENST00000510405.1
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr5_-_114961858 | 0.36 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr2_+_189157536 | 0.35 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_189157498 | 0.33 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr1_+_222988363 | 0.32 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr22_-_29138386 | 0.32 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr8_-_101348408 | 0.32 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr14_+_22600515 | 0.31 |
ENST00000390456.3
|
TRAV8-7
|
T cell receptor alpha variable 8-7 (non-functional) |
| chr17_-_26926005 | 0.31 |
ENST00000536674.2
|
SPAG5
|
sperm associated antigen 5 |
| chr13_+_43597269 | 0.30 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr14_+_54976603 | 0.30 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr15_+_23255242 | 0.28 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr7_+_130794878 | 0.27 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr3_-_101232019 | 0.27 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr4_-_10459009 | 0.26 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr11_-_18610214 | 0.26 |
ENST00000300038.7
ENST00000396197.3 ENST00000320750.6 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr6_+_54711533 | 0.26 |
ENST00000306858.7
|
FAM83B
|
family with sequence similarity 83, member B |
| chr2_-_113191096 | 0.25 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr7_+_97910962 | 0.24 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chr17_-_4463856 | 0.24 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr9_+_114393581 | 0.24 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr1_+_225965518 | 0.24 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr6_-_146285221 | 0.24 |
ENST00000367503.3
ENST00000438092.2 ENST00000275233.7 |
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr6_+_96025341 | 0.24 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chrX_+_51486481 | 0.24 |
ENST00000340438.4
|
GSPT2
|
G1 to S phase transition 2 |
| chr9_-_21995249 | 0.23 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr12_+_102091400 | 0.23 |
ENST00000229266.3
ENST00000549872.1 |
CHPT1
|
choline phosphotransferase 1 |
| chr9_+_86595626 | 0.23 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr3_-_50360165 | 0.22 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr2_-_40006357 | 0.22 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr11_-_2906979 | 0.22 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr10_-_126849626 | 0.21 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr3_+_43732362 | 0.21 |
ENST00000458276.2
|
ABHD5
|
abhydrolase domain containing 5 |
| chr13_+_103451399 | 0.21 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr4_+_38665810 | 0.21 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr1_+_220921582 | 0.21 |
ENST00000359316.2
|
MARC2
|
mitochondrial amidoxime reducing component 2 |
| chr12_+_67663056 | 0.20 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr14_-_45603657 | 0.20 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr15_+_76135622 | 0.20 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr13_-_52026730 | 0.20 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr9_+_114393634 | 0.20 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr12_-_44152551 | 0.20 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr8_+_67344710 | 0.20 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr7_-_105752971 | 0.20 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr14_+_101295948 | 0.19 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr5_+_63461642 | 0.19 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr10_+_115438920 | 0.19 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr2_-_175351744 | 0.19 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr4_-_87855851 | 0.19 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr4_-_76439483 | 0.19 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr4_+_184580420 | 0.19 |
ENST00000334690.6
ENST00000357207.4 |
TRAPPC11
|
trafficking protein particle complex 11 |
| chr1_-_93426998 | 0.18 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr1_-_222763240 | 0.18 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr7_+_30068260 | 0.18 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr1_+_219347203 | 0.18 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr5_+_59783540 | 0.18 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_-_85156417 | 0.18 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr7_-_21985656 | 0.18 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr1_-_152061537 | 0.17 |
ENST00000368806.1
|
TCHHL1
|
trichohyalin-like 1 |
| chr1_-_16939976 | 0.17 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr4_-_184580304 | 0.17 |
ENST00000510968.1
ENST00000512740.1 ENST00000327570.9 |
RWDD4
|
RWD domain containing 4 |
| chr1_-_211848899 | 0.17 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr14_-_71107921 | 0.17 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_+_29241027 | 0.17 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr5_+_118690466 | 0.17 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr10_+_62538248 | 0.17 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr1_+_160709055 | 0.16 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr6_-_110500826 | 0.16 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr1_+_76251912 | 0.16 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr13_+_53030107 | 0.16 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr10_+_60144782 | 0.16 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr11_+_114310237 | 0.16 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr9_-_125027079 | 0.16 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr4_-_159592996 | 0.16 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr13_+_53029564 | 0.16 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr10_+_60145155 | 0.15 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr19_-_40030861 | 0.15 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr12_-_56848426 | 0.15 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr11_-_130184470 | 0.15 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr7_-_100239132 | 0.15 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr18_+_12947981 | 0.15 |
ENST00000262124.11
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr6_+_12008986 | 0.15 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_-_59165763 | 0.15 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chr5_+_112312454 | 0.15 |
ENST00000543319.1
|
DCP2
|
decapping mRNA 2 |
| chr3_-_88108192 | 0.15 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr13_+_25670268 | 0.15 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr16_+_31885079 | 0.15 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr5_+_112312399 | 0.15 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr11_-_8986279 | 0.15 |
ENST00000534025.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr7_-_30544405 | 0.15 |
ENST00000409390.1
ENST00000409144.1 ENST00000005374.6 ENST00000409436.1 ENST00000275428.4 |
GGCT
|
gamma-glutamylcyclotransferase |
| chr6_-_146285455 | 0.14 |
ENST00000367505.2
|
SHPRH
|
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr7_-_105752651 | 0.14 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr2_+_171673072 | 0.14 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr13_+_73356197 | 0.14 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr19_+_44331555 | 0.14 |
ENST00000590950.1
|
ZNF283
|
zinc finger protein 283 |
| chr15_-_66790146 | 0.14 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr11_-_102323489 | 0.14 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr5_+_131892815 | 0.14 |
ENST00000453394.1
|
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr12_+_22778116 | 0.14 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr1_-_40367668 | 0.14 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr2_-_86790472 | 0.14 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr11_-_12030681 | 0.14 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_+_222988406 | 0.14 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr5_-_171433548 | 0.14 |
ENST00000517395.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr11_+_58910201 | 0.14 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr15_+_22736484 | 0.14 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr16_-_88752889 | 0.14 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr2_-_208030886 | 0.13 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr19_+_11877838 | 0.13 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr16_-_14724131 | 0.13 |
ENST00000341484.7
|
PARN
|
poly(A)-specific ribonuclease |
| chr2_+_157291953 | 0.13 |
ENST00000310454.6
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr11_-_18610246 | 0.13 |
ENST00000379387.4
ENST00000541984.1 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr5_-_114961673 | 0.13 |
ENST00000333314.3
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr1_+_109289279 | 0.13 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr16_+_68573640 | 0.13 |
ENST00000398253.2
ENST00000573161.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr9_-_135230336 | 0.13 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr11_+_1856034 | 0.13 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr6_-_57086200 | 0.13 |
ENST00000468148.1
|
RAB23
|
RAB23, member RAS oncogene family |
| chr11_+_105948216 | 0.13 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr6_+_31540056 | 0.13 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr10_+_23728198 | 0.13 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr13_-_48575401 | 0.13 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr6_-_134861089 | 0.13 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr18_-_812517 | 0.12 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr1_-_200379180 | 0.12 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr5_-_55290773 | 0.12 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr1_-_33168336 | 0.12 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr19_+_5904866 | 0.12 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr2_-_122407007 | 0.12 |
ENST00000263710.4
ENST00000455322.2 ENST00000397587.3 ENST00000541377.1 |
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr9_+_139873264 | 0.12 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr22_-_29137771 | 0.12 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr3_+_44596679 | 0.12 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr7_-_16460863 | 0.12 |
ENST00000407010.2
ENST00000399310.3 |
ISPD
|
isoprenoid synthase domain containing |
| chr21_-_34144157 | 0.12 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr4_+_76439665 | 0.12 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr5_+_170288856 | 0.12 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr6_+_133562472 | 0.12 |
ENST00000430974.2
ENST00000367895.5 ENST00000355167.3 ENST00000355286.6 |
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr1_+_91966384 | 0.12 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr1_+_222988464 | 0.12 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr10_+_62538089 | 0.12 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr8_-_74659068 | 0.12 |
ENST00000523558.1
ENST00000521210.1 ENST00000355780.5 ENST00000524104.1 ENST00000521736.1 ENST00000521447.1 ENST00000517542.1 ENST00000521451.1 ENST00000521419.1 ENST00000518502.1 ENST00000524300.1 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr18_-_812231 | 0.12 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr3_-_123680246 | 0.12 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr14_-_64194745 | 0.12 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr11_+_1855645 | 0.11 |
ENST00000381968.3
ENST00000381978.3 |
SYT8
|
synaptotagmin VIII |
| chr2_-_235405679 | 0.11 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr6_-_127664475 | 0.11 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr6_+_71377567 | 0.11 |
ENST00000422334.2
|
SMAP1
|
small ArfGAP 1 |
| chr8_-_494824 | 0.11 |
ENST00000427263.2
ENST00000324079.6 |
TDRP
|
testis development related protein |
| chr18_-_6929829 | 0.11 |
ENST00000583840.1
ENST00000581571.1 ENST00000578497.1 ENST00000579012.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr14_-_23791484 | 0.11 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr7_-_148725544 | 0.11 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr11_-_8985927 | 0.11 |
ENST00000528117.1
ENST00000309134.5 |
TMEM9B
|
TMEM9 domain family, member B |
| chr2_+_197504278 | 0.11 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr1_-_23694794 | 0.11 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr11_-_66445219 | 0.11 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr9_-_86432547 | 0.11 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr15_+_80351977 | 0.11 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_-_107237340 | 0.11 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase |
| chr6_+_111303218 | 0.11 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr1_+_145611010 | 0.11 |
ENST00000369291.5
|
RNF115
|
ring finger protein 115 |
| chr4_+_144106063 | 0.11 |
ENST00000510377.1
|
USP38
|
ubiquitin specific peptidase 38 |
| chr19_+_13122980 | 0.11 |
ENST00000590027.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr19_-_37958318 | 0.11 |
ENST00000316950.6
ENST00000591710.1 |
ZNF569
|
zinc finger protein 569 |
| chr15_+_90777424 | 0.10 |
ENST00000561433.1
ENST00000559204.1 ENST00000558291.1 |
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr2_+_47630108 | 0.10 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr15_+_40544749 | 0.10 |
ENST00000559617.1
ENST00000560684.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr17_+_19314432 | 0.10 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chrX_+_100878079 | 0.10 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr4_+_159593418 | 0.10 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr17_+_48172639 | 0.10 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr20_-_34638841 | 0.10 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr4_+_107236692 | 0.10 |
ENST00000510207.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chrX_-_47863348 | 0.10 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr19_+_16435625 | 0.10 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr11_-_61196858 | 0.10 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr5_-_16509101 | 0.10 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_+_10048831 | 0.10 |
ENST00000581040.1
|
AC000003.2
|
CDNA FLJ25865 fis, clone CBR01927 |
| chr16_+_68279256 | 0.10 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr4_-_107237374 | 0.10 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr9_-_21335240 | 0.10 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr3_+_184529929 | 0.10 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr8_+_70404996 | 0.10 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr19_+_38880695 | 0.10 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_+_32605350 | 0.10 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr4_+_107236847 | 0.10 |
ENST00000358008.3
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr11_-_8986474 | 0.10 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr21_+_34144411 | 0.10 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr16_-_14724057 | 0.09 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr1_-_11918988 | 0.09 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr10_+_94608218 | 0.09 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr6_-_127664683 | 0.09 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr13_-_73356009 | 0.09 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr1_-_202896310 | 0.09 |
ENST00000367261.3
|
KLHL12
|
kelch-like family member 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.5 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.4 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.3 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.2 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.1 | 0.2 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.3 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.1 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.6 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0010520 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.2 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) renal water absorption(GO:0070295) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:1903524 | regulation of vasoconstriction(GO:0019229) vasoconstriction(GO:0042310) positive regulation of blood circulation(GO:1903524) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0015820 | leucine transport(GO:0015820) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 0.2 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.3 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.6 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.2 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.3 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |