Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
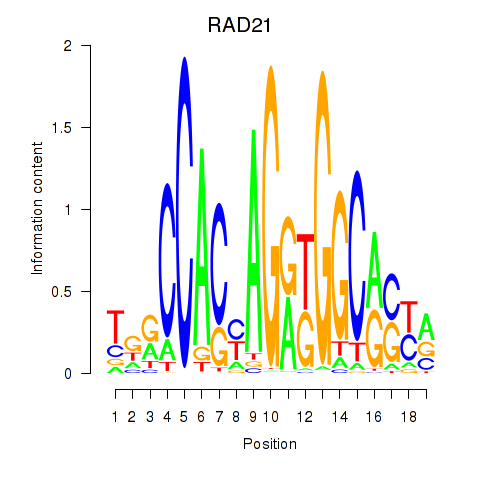
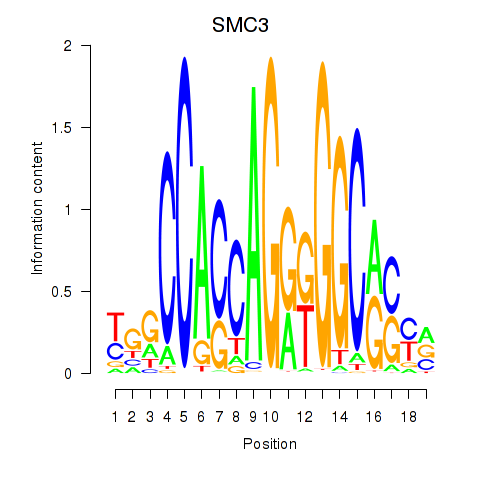
Results for RAD21_SMC3
Z-value: 1.02
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RAD21
|
ENSG00000164754.8 | RAD21 cohesin complex component |
|
SMC3
|
ENSG00000108055.9 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMC3 | hg19_v2_chr10_+_112327425_112327516 | -0.85 | 3.3e-02 | Click! |
| RAD21 | hg19_v2_chr8_-_117886732_117886767 | -0.79 | 6.1e-02 | Click! |
Activity profile of RAD21_SMC3 motif
Sorted Z-values of RAD21_SMC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_45722727 | 1.93 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr2_+_114737472 | 1.41 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr6_+_138188551 | 1.38 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_+_37940153 | 1.30 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr6_+_138188351 | 1.27 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_+_24630465 | 1.20 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr6_+_138188378 | 1.19 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr12_-_7245125 | 1.12 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr5_-_139726181 | 1.10 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr12_-_58159361 | 0.94 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr12_-_7245018 | 0.92 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr18_-_26941900 | 0.91 |
ENST00000577674.1
|
CTD-2515C13.2
|
CTD-2515C13.2 |
| chr2_+_228678550 | 0.89 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr11_+_118826999 | 0.87 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr12_-_7245080 | 0.85 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr10_+_104154229 | 0.79 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr14_+_52118694 | 0.73 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr8_-_11873043 | 0.67 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr2_+_27309605 | 0.66 |
ENST00000260599.6
ENST00000260598.5 ENST00000429697.1 |
KHK
|
ketohexokinase (fructokinase) |
| chr12_-_7245152 | 0.65 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr15_-_38856836 | 0.64 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr9_+_71650645 | 0.60 |
ENST00000396366.2
|
FXN
|
frataxin |
| chr1_-_146989697 | 0.57 |
ENST00000437831.1
|
LINC00624
|
long intergenic non-protein coding RNA 624 |
| chr17_-_48277552 | 0.52 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr15_-_34502197 | 0.52 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr17_+_76356516 | 0.52 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr16_-_22012419 | 0.46 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr7_-_45957011 | 0.45 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr14_-_23292596 | 0.44 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_-_26220366 | 0.43 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr19_-_55652290 | 0.43 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr12_-_82752159 | 0.43 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr1_+_840205 | 0.43 |
ENST00000607769.1
|
RP11-54O7.16
|
RP11-54O7.16 |
| chr8_+_24772455 | 0.39 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr7_-_45956856 | 0.39 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr16_+_66638685 | 0.37 |
ENST00000565003.1
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr9_+_131901710 | 0.36 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr20_+_48807351 | 0.36 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr9_+_116917807 | 0.35 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr11_+_73675873 | 0.34 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr1_+_16767195 | 0.34 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr7_-_6388389 | 0.34 |
ENST00000578372.1
|
FAM220A
|
family with sequence similarity 220, member A |
| chr1_-_63782888 | 0.34 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr9_+_103189458 | 0.33 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr1_+_16767167 | 0.33 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr3_+_9774164 | 0.33 |
ENST00000426583.1
|
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr5_+_172571445 | 0.33 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr1_-_8075693 | 0.32 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr2_-_220025263 | 0.32 |
ENST00000457600.1
|
NHEJ1
|
nonhomologous end-joining factor 1 |
| chr11_+_120039685 | 0.32 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr16_-_90085824 | 0.31 |
ENST00000002501.6
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr12_+_58176525 | 0.31 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr17_+_41561317 | 0.31 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chrX_+_23801280 | 0.31 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr6_-_44225231 | 0.31 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr2_+_113342011 | 0.31 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_+_7591639 | 0.30 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr16_+_22308717 | 0.30 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr1_+_212208919 | 0.30 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr10_+_81107216 | 0.29 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr7_-_5569588 | 0.29 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr20_-_34542548 | 0.29 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr19_-_55653259 | 0.29 |
ENST00000593194.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr1_-_35325400 | 0.29 |
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12 |
| chr6_+_31105426 | 0.29 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr11_-_407103 | 0.28 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr17_+_48556158 | 0.28 |
ENST00000258955.2
|
RSAD1
|
radical S-adenosyl methionine domain containing 1 |
| chr20_-_1447467 | 0.27 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr19_-_49496557 | 0.27 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr19_+_49496782 | 0.27 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr21_+_42741979 | 0.26 |
ENST00000543692.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr17_-_7518145 | 0.26 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr14_-_75079026 | 0.26 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr8_-_11325047 | 0.26 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr15_+_75074410 | 0.26 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr11_+_44117219 | 0.25 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr16_+_2255447 | 0.25 |
ENST00000562352.1
ENST00000562479.1 ENST00000301724.10 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr19_+_45690646 | 0.25 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr17_+_38137073 | 0.25 |
ENST00000541736.1
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr19_+_49496705 | 0.25 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr3_-_50340996 | 0.24 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr16_-_2390704 | 0.24 |
ENST00000301732.5
ENST00000382381.3 |
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr9_+_103189405 | 0.24 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr7_-_99679324 | 0.24 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr22_-_19132154 | 0.24 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr11_+_60691924 | 0.23 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr1_+_28844778 | 0.23 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr17_-_1090599 | 0.23 |
ENST00000544583.2
|
ABR
|
active BCR-related |
| chr2_-_128643496 | 0.23 |
ENST00000272647.5
|
AMMECR1L
|
AMMECR1-like |
| chr9_+_130965651 | 0.23 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr19_-_3547305 | 0.23 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr19_-_36001386 | 0.23 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr12_-_113573495 | 0.22 |
ENST00000446861.3
|
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr10_-_6622258 | 0.22 |
ENST00000263125.5
|
PRKCQ
|
protein kinase C, theta |
| chr16_-_19897455 | 0.22 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chrX_+_49020121 | 0.22 |
ENST00000415364.1
ENST00000376338.3 ENST00000425285.1 |
MAGIX
|
MAGI family member, X-linked |
| chr20_-_1447547 | 0.22 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr20_-_30311703 | 0.22 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr17_-_19265855 | 0.22 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr7_-_27219632 | 0.22 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr16_+_75032901 | 0.21 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr10_-_6622201 | 0.21 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr7_-_91509972 | 0.21 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr4_+_7045042 | 0.21 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr19_-_6481776 | 0.21 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr10_-_49732281 | 0.21 |
ENST00000374170.1
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr10_-_15413035 | 0.21 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr15_-_40212363 | 0.21 |
ENST00000299092.3
|
GPR176
|
G protein-coupled receptor 176 |
| chr22_+_22764088 | 0.21 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr12_-_95510743 | 0.21 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr19_-_1876156 | 0.21 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr11_+_73000449 | 0.20 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr10_+_12391685 | 0.19 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr19_+_16222678 | 0.19 |
ENST00000586682.1
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr17_-_8286484 | 0.19 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr8_-_145550571 | 0.19 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr9_-_38069208 | 0.19 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr1_+_20208870 | 0.19 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr8_+_42948641 | 0.19 |
ENST00000518991.1
ENST00000331373.5 |
POMK
|
protein-O-mannose kinase |
| chr12_-_51663728 | 0.19 |
ENST00000603864.1
ENST00000605426.1 |
SMAGP
|
small cell adhesion glycoprotein |
| chr18_-_47721447 | 0.19 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr12_+_51985001 | 0.19 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr16_+_2867164 | 0.19 |
ENST00000455114.1
ENST00000450020.3 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr17_+_7591747 | 0.19 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr19_+_49977466 | 0.19 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr12_-_113574028 | 0.19 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr17_-_33469299 | 0.18 |
ENST00000586869.1
ENST00000360831.5 ENST00000442241.4 |
NLE1
|
notchless homolog 1 (Drosophila) |
| chr11_+_46368956 | 0.18 |
ENST00000543978.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr11_+_46369077 | 0.18 |
ENST00000456247.2
ENST00000421244.2 ENST00000318201.8 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_-_113249678 | 0.18 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr9_-_94712434 | 0.18 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr17_-_19265982 | 0.18 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr11_+_46368975 | 0.18 |
ENST00000527911.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr22_-_30783075 | 0.17 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr17_+_38137050 | 0.17 |
ENST00000264639.4
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr5_-_140998616 | 0.17 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr10_-_6019984 | 0.17 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr1_+_154244987 | 0.17 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr1_+_145524891 | 0.17 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr1_-_6761855 | 0.17 |
ENST00000426784.1
ENST00000377573.5 ENST00000377577.5 ENST00000294401.7 |
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr9_-_132404374 | 0.17 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr4_-_171011084 | 0.17 |
ENST00000337664.4
|
AADAT
|
aminoadipate aminotransferase |
| chr17_+_1958388 | 0.17 |
ENST00000399849.3
|
HIC1
|
hypermethylated in cancer 1 |
| chrX_+_49969405 | 0.17 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr10_-_76995769 | 0.17 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr22_-_22307199 | 0.16 |
ENST00000397495.4
ENST00000263212.5 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr2_+_234263120 | 0.16 |
ENST00000264057.2
ENST00000427930.1 |
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr10_+_35416223 | 0.16 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chrX_-_27417088 | 0.16 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr9_+_34646651 | 0.16 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr12_-_89918982 | 0.16 |
ENST00000549504.1
|
POC1B
|
POC1 centriolar protein B |
| chr16_-_30441293 | 0.16 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr16_+_2255710 | 0.16 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chrX_+_47053208 | 0.16 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr3_-_190040223 | 0.16 |
ENST00000295522.3
|
CLDN1
|
claudin 1 |
| chr10_+_17272608 | 0.16 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr5_-_146833222 | 0.15 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr16_+_2867228 | 0.15 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr11_-_117102768 | 0.15 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr5_+_148521381 | 0.15 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr2_+_171785824 | 0.15 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr6_-_32920794 | 0.15 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr6_+_35310312 | 0.15 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr16_+_15737124 | 0.15 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr19_+_16222439 | 0.15 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr19_+_4791722 | 0.15 |
ENST00000269856.3
|
FEM1A
|
fem-1 homolog a (C. elegans) |
| chr9_+_103189660 | 0.15 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr8_+_67341239 | 0.15 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr16_+_69345243 | 0.15 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr16_-_69788816 | 0.15 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr11_+_61520075 | 0.15 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr2_+_171785012 | 0.14 |
ENST00000234160.4
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr17_+_65375082 | 0.14 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr6_+_151186554 | 0.14 |
ENST00000367321.3
ENST00000367307.4 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_+_196440692 | 0.14 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_+_34646624 | 0.14 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr16_+_55357672 | 0.14 |
ENST00000290552.7
|
IRX6
|
iroquois homeobox 6 |
| chr9_+_129622904 | 0.14 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chrX_-_46618490 | 0.14 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr19_-_36001286 | 0.14 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr9_+_130965677 | 0.14 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr16_-_49315731 | 0.14 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr4_+_56814968 | 0.14 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr11_+_62554860 | 0.14 |
ENST00000533861.1
ENST00000333449.4 |
TMEM179B
|
transmembrane protein 179B |
| chr17_+_73629500 | 0.14 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr22_+_31160239 | 0.13 |
ENST00000445781.1
ENST00000401475.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr2_+_131113580 | 0.13 |
ENST00000175756.5
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr1_-_113249734 | 0.13 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
| chr1_+_64239657 | 0.13 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr7_+_89783689 | 0.13 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr16_+_89642120 | 0.13 |
ENST00000268720.5
ENST00000319518.8 |
CPNE7
|
copine VII |
| chr3_-_128369643 | 0.13 |
ENST00000296255.3
|
RPN1
|
ribophorin I |
| chr13_-_88323514 | 0.13 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr11_-_117103208 | 0.13 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr15_+_75074385 | 0.13 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr5_-_140998481 | 0.13 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr5_+_148521046 | 0.13 |
ENST00000326685.7
ENST00000356541.3 ENST00000309868.7 |
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr15_-_71407806 | 0.13 |
ENST00000566432.1
ENST00000567117.1 |
CT62
|
cancer/testis antigen 62 |
| chr14_+_101293687 | 0.13 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chrX_-_102942961 | 0.13 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RAD21_SMC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.4 | 1.3 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.3 | 0.9 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 0.9 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.8 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 0.5 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 0.6 | GO:0019230 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) proprioception(GO:0019230) |
| 0.1 | 0.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 1.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.7 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.4 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.6 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.5 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.3 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.1 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 3.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.4 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.3 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.6 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0035106 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0044843 | cell cycle G1/S phase transition(GO:0044843) |
| 0.0 | 0.1 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 1.3 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:1903027 | asymmetric Golgi ribbon formation(GO:0090164) regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.0 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.0 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 0.8 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 1.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.5 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 0.8 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.2 | 3.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 4.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |