Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
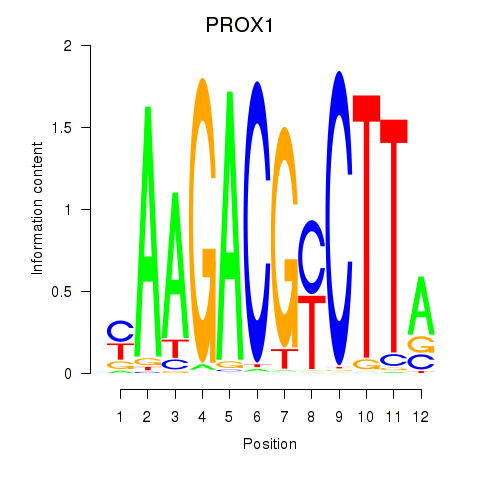
Results for PROX1
Z-value: 0.68
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161854_214161892 | -0.70 | 1.2e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_14920599 | 0.39 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr1_+_209545365 | 0.30 |
ENST00000447257.1
|
RP11-372M18.2
|
RP11-372M18.2 |
| chr18_-_2571210 | 0.28 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr19_+_49535169 | 0.27 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr7_-_143582450 | 0.27 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr4_-_48683188 | 0.26 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr3_+_49941420 | 0.26 |
ENST00000419183.1
|
CTD-2330K9.3
|
Uncharacterized protein |
| chr19_-_5293243 | 0.24 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr3_-_96337000 | 0.23 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr21_+_35736302 | 0.21 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr4_+_128886584 | 0.20 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr19_-_53324884 | 0.19 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr19_+_58281014 | 0.19 |
ENST00000391702.3
ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586
|
zinc finger protein 586 |
| chr1_+_104068562 | 0.18 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr2_+_163175394 | 0.18 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr10_+_14920843 | 0.18 |
ENST00000433779.1
ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr14_-_35183755 | 0.17 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr12_+_42624050 | 0.17 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr20_-_61885826 | 0.16 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr16_+_85832146 | 0.15 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr10_-_92681033 | 0.15 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr17_+_66539369 | 0.15 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr6_-_86303833 | 0.14 |
ENST00000505648.1
|
SNX14
|
sorting nexin 14 |
| chr3_-_25915189 | 0.14 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr17_+_16593539 | 0.14 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr16_+_28857916 | 0.14 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr7_+_64254793 | 0.13 |
ENST00000494380.1
ENST00000440155.2 ENST00000440598.1 ENST00000437743.1 |
ZNF138
|
zinc finger protein 138 |
| chr9_+_125133315 | 0.13 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_73629500 | 0.13 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr11_-_82997371 | 0.13 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr1_+_104068312 | 0.13 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr4_-_103747011 | 0.13 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_27384737 | 0.13 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr3_-_197024965 | 0.12 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_53073526 | 0.12 |
ENST00000596514.1
ENST00000391785.3 ENST00000301093.2 |
ZNF701
|
zinc finger protein 701 |
| chr2_-_107084826 | 0.12 |
ENST00000304514.7
ENST00000409886.3 |
RGPD3
|
RANBP2-like and GRIP domain containing 3 |
| chr11_-_61062762 | 0.12 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr2_+_108443388 | 0.12 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr11_-_111749767 | 0.12 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr6_-_107235378 | 0.11 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr16_+_31885079 | 0.11 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr2_-_54087066 | 0.11 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr9_+_72873837 | 0.11 |
ENST00000361138.5
|
SMC5
|
structural maintenance of chromosomes 5 |
| chr22_+_24990746 | 0.11 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr19_+_15752088 | 0.11 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr18_+_2571510 | 0.11 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr6_+_90192974 | 0.11 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr1_-_154178803 | 0.11 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr2_+_29341037 | 0.10 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr1_+_241695424 | 0.10 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_+_57618269 | 0.10 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr4_-_169931393 | 0.10 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr3_-_197025447 | 0.10 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr7_+_64254766 | 0.10 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr15_+_38746307 | 0.10 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr5_-_156569754 | 0.09 |
ENST00000420343.1
|
MED7
|
mediator complex subunit 7 |
| chr2_-_225434538 | 0.09 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr9_+_125132803 | 0.09 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr16_+_23194033 | 0.09 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr3_-_196065248 | 0.09 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr6_-_28226984 | 0.09 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr10_-_94301107 | 0.09 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr5_+_64859066 | 0.09 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr12_-_46766577 | 0.09 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr21_-_34863693 | 0.09 |
ENST00000314399.3
|
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr3_-_137834436 | 0.09 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr5_+_6766004 | 0.09 |
ENST00000506093.1
|
RP11-332J15.3
|
RP11-332J15.3 |
| chr4_-_170947522 | 0.09 |
ENST00000361618.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_-_159592996 | 0.09 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr16_-_21442874 | 0.09 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr21_-_16125773 | 0.09 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr11_-_111749878 | 0.09 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr2_+_241499719 | 0.08 |
ENST00000405954.1
|
DUSP28
|
dual specificity phosphatase 28 |
| chr10_+_60028818 | 0.08 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr11_+_28131821 | 0.08 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr6_+_111580508 | 0.08 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr1_-_207226313 | 0.08 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr1_+_95975672 | 0.08 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr16_-_23607598 | 0.08 |
ENST00000562133.1
ENST00000570319.1 ENST00000007516.3 |
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
| chr15_-_78913628 | 0.08 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr15_+_81489213 | 0.08 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr4_+_26862313 | 0.08 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chr17_+_38337491 | 0.08 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr3_+_179040767 | 0.08 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr4_-_169931231 | 0.08 |
ENST00000504561.1
|
CBR4
|
carbonyl reductase 4 |
| chr4_+_2819883 | 0.08 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_+_9839335 | 0.08 |
ENST00000453882.1
|
ARPC4-TTLL3
|
ARPC4-TTLL3 readthrough |
| chr10_-_123357598 | 0.08 |
ENST00000358487.5
ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2
|
fibroblast growth factor receptor 2 |
| chr4_+_26862400 | 0.07 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr16_-_22012419 | 0.07 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr11_+_125464758 | 0.07 |
ENST00000529886.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr6_+_117996621 | 0.07 |
ENST00000368494.3
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
| chr20_-_749121 | 0.07 |
ENST00000217254.7
ENST00000381944.3 |
SLC52A3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr6_+_170151887 | 0.07 |
ENST00000588451.1
|
ERMARD
|
ER membrane-associated RNA degradation |
| chr6_+_170151708 | 0.07 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr5_+_177433973 | 0.07 |
ENST00000507848.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr3_-_18487057 | 0.07 |
ENST00000415069.1
|
SATB1
|
SATB homeobox 1 |
| chr10_+_16478942 | 0.07 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr7_-_102257139 | 0.07 |
ENST00000521076.1
ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4
|
RAS p21 protein activator 4 |
| chr3_-_27410847 | 0.07 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr10_-_96122682 | 0.07 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr4_+_26862431 | 0.06 |
ENST00000465503.1
|
STIM2
|
stromal interaction molecule 2 |
| chr1_+_246729724 | 0.06 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr16_+_22518495 | 0.06 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_+_31724552 | 0.06 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr5_+_131892603 | 0.06 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr6_-_74104856 | 0.06 |
ENST00000441145.1
|
OOEP
|
oocyte expressed protein |
| chr14_+_37641012 | 0.06 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr1_+_35544968 | 0.06 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr7_-_156433195 | 0.06 |
ENST00000333319.6
|
C7orf13
|
chromosome 7 open reading frame 13 |
| chrX_+_117861535 | 0.06 |
ENST00000371666.3
ENST00000371642.1 |
IL13RA1
|
interleukin 13 receptor, alpha 1 |
| chr3_+_152552685 | 0.06 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr12_-_122296755 | 0.06 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chrX_+_150565038 | 0.06 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr21_-_16126181 | 0.06 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr5_-_156569850 | 0.06 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr9_-_130497565 | 0.06 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr3_+_138067314 | 0.06 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr5_-_177210399 | 0.06 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr4_-_69215467 | 0.05 |
ENST00000579690.1
|
YTHDC1
|
YTH domain containing 1 |
| chr2_+_28718921 | 0.05 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr11_+_117947724 | 0.05 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr5_-_180688105 | 0.05 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr4_-_170947446 | 0.05 |
ENST00000507601.1
ENST00000512698.1 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr17_+_26646121 | 0.05 |
ENST00000226230.6
|
TMEM97
|
transmembrane protein 97 |
| chr6_-_159420780 | 0.05 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr2_-_42588338 | 0.05 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr8_+_104310661 | 0.05 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr6_-_134495992 | 0.05 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_-_9546227 | 0.05 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr12_+_52668394 | 0.05 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr14_-_54425475 | 0.05 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr8_+_110552046 | 0.05 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr12_-_51418549 | 0.05 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr4_-_103746924 | 0.05 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103746683 | 0.05 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_66093851 | 0.05 |
ENST00000275532.3
|
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr10_+_112327425 | 0.05 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr6_+_126102292 | 0.05 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_-_73882057 | 0.04 |
ENST00000334126.7
ENST00000313663.7 |
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr8_-_143696833 | 0.04 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr4_-_170947485 | 0.04 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr17_+_66243715 | 0.04 |
ENST00000359904.3
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr14_-_50559361 | 0.04 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr1_+_150980989 | 0.04 |
ENST00000368935.1
|
PRUNE
|
prune exopolyphosphatase |
| chr11_-_118023490 | 0.04 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr11_+_61522844 | 0.04 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr16_-_12070468 | 0.04 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr15_-_90892669 | 0.04 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr19_+_13858701 | 0.04 |
ENST00000586666.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr18_-_21562500 | 0.04 |
ENST00000582300.2
|
RP11-403A21.1
|
RP11-403A21.1 |
| chr19_-_46234119 | 0.04 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr11_+_61722629 | 0.04 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chr1_-_44497118 | 0.04 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_-_86571628 | 0.04 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr10_-_52008313 | 0.04 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr6_-_35992270 | 0.04 |
ENST00000394602.2
ENST00000355574.2 |
SLC26A8
|
solute carrier family 26 (anion exchanger), member 8 |
| chr1_+_26759295 | 0.04 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr12_-_122240792 | 0.04 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr2_-_233877912 | 0.04 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr22_-_29075853 | 0.04 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr14_-_31676964 | 0.04 |
ENST00000553700.1
|
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr7_-_91509986 | 0.04 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr1_+_241695670 | 0.04 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_-_73882249 | 0.04 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr8_-_41166953 | 0.04 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr12_-_117628253 | 0.04 |
ENST00000330622.5
|
FBXO21
|
F-box protein 21 |
| chr3_-_196014520 | 0.04 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr7_+_100209979 | 0.03 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr1_-_28241226 | 0.03 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr6_-_137540477 | 0.03 |
ENST00000367735.2
ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chr11_+_93394805 | 0.03 |
ENST00000325212.6
ENST00000411936.1 ENST00000344196.4 |
KIAA1731
|
KIAA1731 |
| chr1_-_109203685 | 0.03 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr21_-_26979786 | 0.03 |
ENST00000419219.1
ENST00000352957.4 ENST00000307301.7 |
MRPL39
|
mitochondrial ribosomal protein L39 |
| chr14_-_23426337 | 0.03 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr16_+_57279248 | 0.03 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr1_-_118727781 | 0.03 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr17_-_36904437 | 0.03 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr5_+_175490540 | 0.03 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr1_-_152539248 | 0.03 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr19_-_19754354 | 0.03 |
ENST00000587238.1
|
GMIP
|
GEM interacting protein |
| chr21_-_40033618 | 0.03 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr14_-_23426322 | 0.03 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr7_+_1609765 | 0.03 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr3_+_186915274 | 0.03 |
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1 |
| chr3_+_136676707 | 0.03 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr14_+_78266408 | 0.03 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr5_+_156693159 | 0.03 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr11_+_57308979 | 0.03 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr10_+_54074033 | 0.03 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr14_-_23426270 | 0.03 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr4_-_128887069 | 0.03 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr7_-_91509972 | 0.02 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr6_-_4079334 | 0.02 |
ENST00000492651.1
ENST00000498677.1 ENST00000274673.3 |
FAM217A
|
family with sequence similarity 217, member A |
| chr9_+_95858485 | 0.02 |
ENST00000375464.2
|
C9orf89
|
chromosome 9 open reading frame 89 |
| chr18_+_29769978 | 0.02 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr14_+_45605157 | 0.02 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr7_+_48128194 | 0.02 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr4_-_170947565 | 0.02 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_109399682 | 0.02 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0060615 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) regulation of penile erection(GO:0060405) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.0 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |