Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
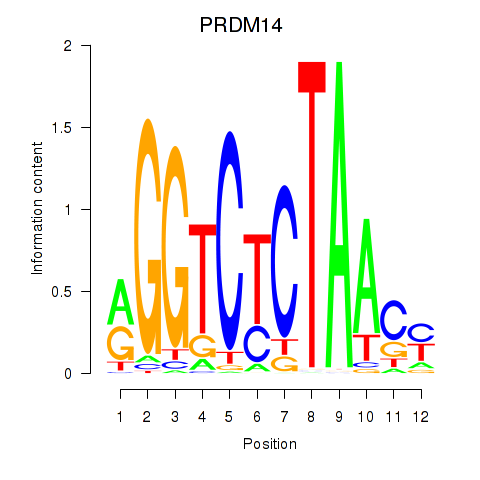
Results for PRDM14
Z-value: 0.53
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | 0.70 | 1.2e-01 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_156052354 | 0.28 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr18_+_57567180 | 0.27 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr13_-_34250861 | 0.27 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr1_+_27668505 | 0.26 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr15_-_45694380 | 0.26 |
ENST00000561148.1
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr4_-_10459009 | 0.24 |
ENST00000507515.1
|
ZNF518B
|
zinc finger protein 518B |
| chr3_-_143567262 | 0.24 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr4_-_103266626 | 0.20 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr12_-_76742183 | 0.20 |
ENST00000393262.3
|
BBS10
|
Bardet-Biedl syndrome 10 |
| chr17_-_19651598 | 0.20 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_134092561 | 0.20 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr17_-_19651654 | 0.19 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_-_116383322 | 0.18 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr12_-_52967600 | 0.15 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr1_-_33642151 | 0.14 |
ENST00000543586.1
|
TRIM62
|
tripartite motif containing 62 |
| chr15_+_63188009 | 0.14 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr4_+_141677577 | 0.13 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr17_-_17726907 | 0.11 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr12_+_49760639 | 0.11 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr20_-_3154162 | 0.11 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr1_+_36621529 | 0.10 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_+_36873677 | 0.10 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr5_-_108063949 | 0.10 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr11_+_85566422 | 0.10 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr12_-_56326402 | 0.10 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr17_-_19651668 | 0.09 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr19_-_6279932 | 0.09 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr8_-_102216925 | 0.08 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr5_-_55777586 | 0.08 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr12_+_121647868 | 0.08 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr3_-_47517302 | 0.07 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr14_-_21492113 | 0.07 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr2_+_101437487 | 0.07 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_-_208030886 | 0.07 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_3600356 | 0.06 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr17_-_73775839 | 0.05 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr1_+_45274154 | 0.05 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr19_+_49838653 | 0.05 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr1_+_36621174 | 0.04 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr6_+_43140095 | 0.04 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr4_+_81187753 | 0.04 |
ENST00000312465.7
ENST00000456523.3 |
FGF5
|
fibroblast growth factor 5 |
| chr11_+_64879317 | 0.04 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr17_-_2117600 | 0.04 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr4_+_4861385 | 0.04 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr7_-_124405681 | 0.04 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr15_+_45694523 | 0.03 |
ENST00000305560.6
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chrX_+_41193407 | 0.03 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr10_+_12171636 | 0.03 |
ENST00000379051.1
ENST00000379033.3 ENST00000441368.1 ENST00000298428.9 ENST00000304267.8 |
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr2_-_192711968 | 0.03 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr11_-_62474803 | 0.03 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr7_-_30066233 | 0.03 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr17_+_41924536 | 0.02 |
ENST00000317310.4
|
CD300LG
|
CD300 molecule-like family member g |
| chr6_-_46048116 | 0.02 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr14_+_89290965 | 0.02 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr10_+_12171721 | 0.02 |
ENST00000379020.4
ENST00000379017.3 |
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr19_-_49314269 | 0.02 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr12_+_72332618 | 0.02 |
ENST00000333850.3
|
TPH2
|
tryptophan hydroxylase 2 |
| chr6_-_109777128 | 0.02 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr14_-_21492251 | 0.02 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr12_+_121647962 | 0.02 |
ENST00000542067.1
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr22_-_29457832 | 0.02 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr15_+_45694567 | 0.01 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr17_-_46691990 | 0.01 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr19_+_3572758 | 0.01 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_47653178 | 0.01 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr9_+_132099158 | 0.01 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr1_-_27216729 | 0.00 |
ENST00000431781.2
ENST00000374135.4 |
GPN2
|
GPN-loop GTPase 2 |
| chr3_-_155394099 | 0.00 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr7_+_39989611 | 0.00 |
ENST00000181839.4
|
CDK13
|
cyclin-dependent kinase 13 |
| chr19_-_5624057 | 0.00 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr19_-_49314169 | 0.00 |
ENST00000597011.1
ENST00000601681.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr6_+_72926145 | 0.00 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |