Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
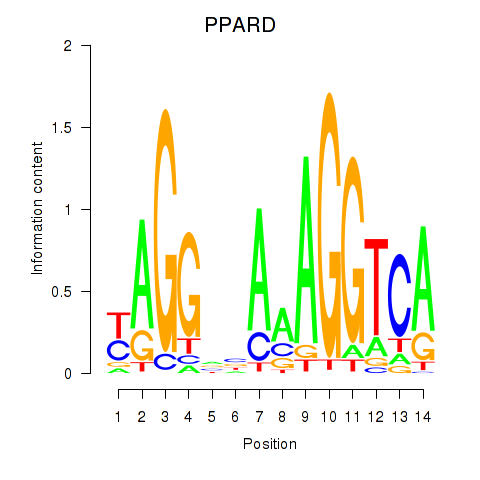
Results for PPARD
Z-value: 0.39
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310391_35310410 | 0.12 | 8.1e-01 | Click! |
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_172484377 | 0.34 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chrX_-_108868390 | 0.24 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr1_+_6511651 | 0.17 |
ENST00000434576.1
|
ESPN
|
espin |
| chr7_-_34978980 | 0.17 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr11_+_66624527 | 0.17 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_111169565 | 0.16 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr15_+_82555125 | 0.15 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr14_+_32546145 | 0.14 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_+_159651821 | 0.14 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr12_+_121163602 | 0.13 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr12_+_121163538 | 0.13 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr6_+_33172407 | 0.12 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr2_+_228190066 | 0.12 |
ENST00000436237.1
ENST00000443428.2 ENST00000418961.1 |
MFF
|
mitochondrial fission factor |
| chr17_+_48610074 | 0.12 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr4_+_159593418 | 0.11 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr14_+_32546274 | 0.11 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr14_+_74551650 | 0.11 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr3_+_48507210 | 0.10 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr17_+_7123207 | 0.10 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr12_-_109797249 | 0.09 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr2_-_28113217 | 0.08 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr11_+_111169976 | 0.08 |
ENST00000398035.2
|
COLCA2
|
colorectal cancer associated 2 |
| chr4_+_159593271 | 0.08 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr2_+_198365122 | 0.08 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr11_-_45939374 | 0.08 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr2_+_178257372 | 0.08 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr2_+_219433588 | 0.08 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr1_+_202317815 | 0.07 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_+_94050913 | 0.07 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr2_-_178257401 | 0.07 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr12_-_95467267 | 0.07 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr5_-_16509101 | 0.07 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr3_-_50605077 | 0.07 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr9_+_131901661 | 0.06 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_-_49339080 | 0.05 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_+_244998602 | 0.05 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr5_+_14664762 | 0.05 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr17_-_4464081 | 0.05 |
ENST00000574154.1
|
GGT6
|
gamma-glutamyltransferase 6 |
| chr12_-_56727676 | 0.05 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr9_+_115142217 | 0.05 |
ENST00000398805.3
ENST00000398803.1 ENST00000262542.7 ENST00000539114.1 |
HSDL2
|
hydroxysteroid dehydrogenase like 2 |
| chr6_+_143999072 | 0.05 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_+_156123318 | 0.05 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr8_-_116681686 | 0.04 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_-_49314640 | 0.04 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr8_+_145065705 | 0.04 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr2_-_99871570 | 0.04 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr6_+_88299833 | 0.04 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr10_-_94050820 | 0.04 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_219283815 | 0.04 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr17_+_4802713 | 0.04 |
ENST00000521575.1
ENST00000381365.3 |
C17orf107
|
chromosome 17 open reading frame 107 |
| chr9_+_35042205 | 0.04 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr20_-_44485835 | 0.04 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr4_+_174089904 | 0.04 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr19_-_3062465 | 0.04 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr6_+_142468383 | 0.03 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr17_+_48610042 | 0.03 |
ENST00000503246.1
|
EPN3
|
epsin 3 |
| chr8_+_145065521 | 0.03 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_+_17281900 | 0.03 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr3_+_186383741 | 0.03 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr12_-_6665200 | 0.03 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr4_-_108204904 | 0.03 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr2_+_219433281 | 0.03 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr15_-_43865633 | 0.03 |
ENST00000437065.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr20_+_46130601 | 0.03 |
ENST00000341724.6
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr19_+_29493456 | 0.02 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr15_+_82555169 | 0.02 |
ENST00000565432.1
ENST00000427381.2 |
FAM154B
|
family with sequence similarity 154, member B |
| chr2_+_95940186 | 0.02 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr4_-_108204846 | 0.02 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr20_+_42187682 | 0.02 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr20_+_19870167 | 0.02 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr1_+_202317855 | 0.02 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_-_160253998 | 0.02 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr2_+_39117010 | 0.02 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr12_-_109219937 | 0.02 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr15_+_43885252 | 0.01 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr20_+_32077880 | 0.01 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr16_-_75285380 | 0.01 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr1_+_62417957 | 0.01 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr21_+_27107672 | 0.01 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr2_+_120125245 | 0.01 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr16_+_2255841 | 0.01 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr15_+_91449971 | 0.01 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr12_-_53320245 | 0.01 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr1_-_11863171 | 0.01 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr6_+_142468361 | 0.01 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr20_+_42187608 | 0.01 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr7_+_7222233 | 0.01 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr7_+_100663353 | 0.01 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chr15_-_82555000 | 0.01 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr1_-_11107280 | 0.01 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr15_+_43985084 | 0.01 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr12_-_56727487 | 0.01 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_-_161102367 | 0.01 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr12_+_96252706 | 0.01 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_+_7222157 | 0.01 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr2_-_219433014 | 0.01 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr2_-_198364581 | 0.01 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr11_-_47470682 | 0.01 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr8_-_80942467 | 0.01 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr11_-_47470591 | 0.01 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr11_-_119599794 | 0.00 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr6_+_143999185 | 0.00 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr5_-_143550159 | 0.00 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr21_-_27107881 | 0.00 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_+_31637944 | 0.00 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr16_+_81272287 | 0.00 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr21_-_27107283 | 0.00 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr19_+_10197463 | 0.00 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_-_29508321 | 0.00 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr3_-_125094093 | 0.00 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr6_-_33547975 | 0.00 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr22_-_29784519 | 0.00 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr11_-_45939565 | 0.00 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr6_-_88299678 | 0.00 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr11_-_47470703 | 0.00 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |