Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
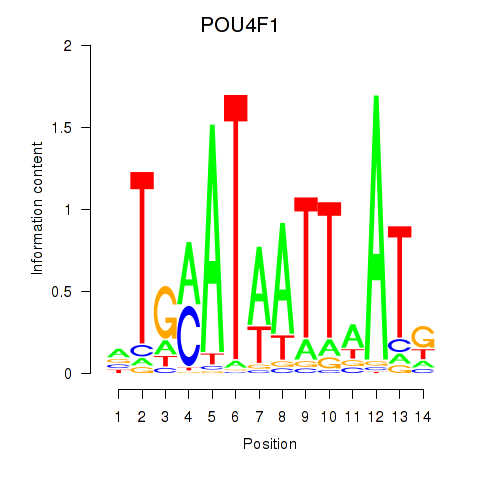
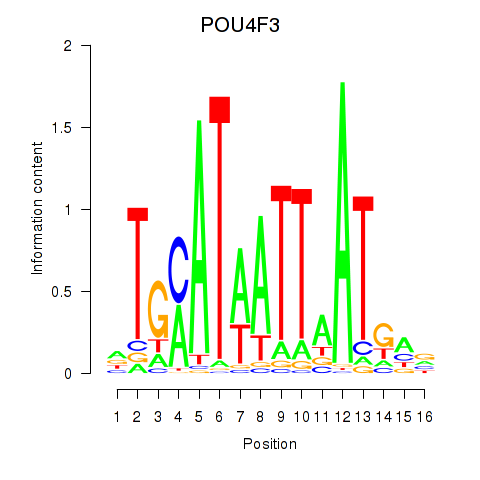
Results for POU4F1_POU4F3
Z-value: 0.31
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | 0.71 | 1.1e-01 | Click! |
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | 0.40 | 4.3e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26108355 | 0.27 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr2_-_55237484 | 0.24 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr6_-_116833500 | 0.21 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr7_+_150725510 | 0.14 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr4_+_141264597 | 0.14 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr19_+_48949087 | 0.13 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr19_-_18632861 | 0.13 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr3_+_167582561 | 0.12 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr12_-_58212487 | 0.12 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr2_+_61372226 | 0.12 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr2_+_179318295 | 0.12 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr19_-_20844343 | 0.11 |
ENST00000595405.1
|
ZNF626
|
zinc finger protein 626 |
| chr6_-_52859046 | 0.10 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr19_+_19639670 | 0.10 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr17_+_17685422 | 0.10 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr19_+_19639704 | 0.10 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr12_-_100486668 | 0.09 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr7_-_35013217 | 0.08 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr22_-_38506619 | 0.08 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr3_-_52860850 | 0.08 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr7_+_134528635 | 0.07 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr1_+_149239529 | 0.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chrX_-_138724677 | 0.07 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr14_+_101297740 | 0.07 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr2_+_179317994 | 0.07 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr11_-_26593677 | 0.07 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr7_-_5465045 | 0.07 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chrX_+_47053208 | 0.07 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr3_+_46742823 | 0.06 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chrX_+_78003204 | 0.06 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr4_+_154622652 | 0.06 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr2_+_183582774 | 0.06 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr20_+_30102231 | 0.06 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr14_-_54908043 | 0.06 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr16_-_70835034 | 0.05 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr14_+_39703112 | 0.05 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr10_+_118349920 | 0.05 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr18_+_61575200 | 0.05 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr19_-_55677999 | 0.05 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr1_+_145292806 | 0.05 |
ENST00000448873.2
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr3_-_33686925 | 0.05 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_-_88285309 | 0.05 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr6_+_127898312 | 0.05 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr12_-_89746264 | 0.05 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_+_36317830 | 0.05 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr2_+_87135076 | 0.05 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr6_+_12717892 | 0.05 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr15_-_42343388 | 0.05 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr3_-_143567262 | 0.05 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr12_+_41221794 | 0.05 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr22_+_39348723 | 0.05 |
ENST00000402255.1
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr10_-_115904361 | 0.04 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr6_-_10115007 | 0.04 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr19_-_23941639 | 0.04 |
ENST00000395385.3
ENST00000531570.1 ENST00000528059.1 |
ZNF681
|
zinc finger protein 681 |
| chr14_+_57671888 | 0.04 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr15_-_54267147 | 0.04 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr10_-_6104253 | 0.04 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chrX_-_1331527 | 0.04 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr7_-_22234381 | 0.04 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr3_-_33686743 | 0.04 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_86085778 | 0.04 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr5_+_68860949 | 0.04 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr19_+_39138271 | 0.04 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chr22_-_42739533 | 0.04 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr7_-_100425112 | 0.04 |
ENST00000358173.3
|
EPHB4
|
EPH receptor B4 |
| chr1_+_81771806 | 0.04 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chrX_-_138724994 | 0.04 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr19_-_55677920 | 0.04 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr6_-_33297013 | 0.03 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr19_-_22018966 | 0.03 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr19_-_20748541 | 0.03 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr11_-_26593649 | 0.03 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_559377 | 0.03 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr2_-_183387064 | 0.03 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_40540586 | 0.03 |
ENST00000264657.5
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr12_+_28410128 | 0.03 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr3_+_141043050 | 0.03 |
ENST00000509842.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_191221240 | 0.03 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr13_+_52586517 | 0.03 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr15_-_65426174 | 0.03 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr17_-_36358166 | 0.03 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr4_-_122085469 | 0.03 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_-_222014008 | 0.03 |
ENST00000431729.1
|
RP11-191N8.2
|
RP11-191N8.2 |
| chr19_+_17865011 | 0.03 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr12_+_109490370 | 0.03 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr4_-_89442940 | 0.03 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr9_-_123812542 | 0.03 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr6_+_34204642 | 0.03 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr12_+_122880045 | 0.03 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr6_+_30856507 | 0.03 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr19_+_39138320 | 0.03 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr5_+_61874562 | 0.03 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr9_-_3469181 | 0.03 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr21_+_45937497 | 0.02 |
ENST00000354333.5
|
C21orf90
|
chromosome 21 open reading frame 90 |
| chr16_+_619931 | 0.02 |
ENST00000321878.5
ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chrX_+_41583408 | 0.02 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr10_+_104614008 | 0.02 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr1_+_73771844 | 0.02 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr17_+_7155819 | 0.02 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_+_133174147 | 0.02 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chr8_-_125577940 | 0.02 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr14_+_39703084 | 0.02 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr12_-_51611477 | 0.02 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr7_-_20256965 | 0.02 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr5_+_129083772 | 0.02 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr18_-_33647487 | 0.02 |
ENST00000590898.1
ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr9_-_85882145 | 0.02 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr2_+_65663812 | 0.02 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr4_+_106631966 | 0.02 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr15_-_52587945 | 0.02 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr12_+_110011571 | 0.02 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr13_+_76413852 | 0.02 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr17_-_29641084 | 0.02 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_+_198608146 | 0.02 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_161695042 | 0.02 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr7_-_115670792 | 0.02 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr2_-_3504587 | 0.02 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr12_-_89746173 | 0.01 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_+_165864821 | 0.01 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_-_95538492 | 0.01 |
ENST00000370205.5
|
ALG14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr17_-_49124230 | 0.01 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr10_+_81891416 | 0.01 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr10_+_112679301 | 0.01 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr2_+_87144738 | 0.01 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr7_-_115670804 | 0.01 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr14_-_65409502 | 0.01 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr5_-_135290705 | 0.01 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr6_-_136847610 | 0.01 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr16_-_85784557 | 0.01 |
ENST00000602675.1
|
C16orf74
|
chromosome 16 open reading frame 74 |
| chr1_+_161691353 | 0.01 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr10_-_11574274 | 0.01 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr14_+_31959154 | 0.01 |
ENST00000550005.1
|
NUBPL
|
nucleotide binding protein-like |
| chr16_-_67217844 | 0.01 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr8_-_101733794 | 0.01 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_8378140 | 0.01 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr2_+_161993465 | 0.01 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_111649015 | 0.01 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_+_130569429 | 0.01 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr17_+_29664830 | 0.01 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr12_+_28605426 | 0.01 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_54303934 | 0.01 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr14_+_31494841 | 0.01 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr6_+_126102292 | 0.01 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_143929307 | 0.01 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr4_+_74269956 | 0.01 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr9_-_4666421 | 0.01 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr18_+_29027696 | 0.01 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr8_+_92261516 | 0.01 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr10_+_33271469 | 0.01 |
ENST00000414157.1
|
RP11-462L8.1
|
RP11-462L8.1 |
| chr16_-_11363178 | 0.01 |
ENST00000312693.3
|
TNP2
|
transition protein 2 (during histone to protamine replacement) |
| chr10_+_104613980 | 0.01 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr2_+_187558698 | 0.01 |
ENST00000304698.5
|
FAM171B
|
family with sequence similarity 171, member B |
| chr5_-_77844974 | 0.01 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr6_-_127840453 | 0.01 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr4_-_68749699 | 0.01 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr6_-_127840336 | 0.01 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr5_-_58295712 | 0.01 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_-_74283694 | 0.01 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_+_161993412 | 0.01 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_40540484 | 0.01 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr10_-_75226166 | 0.01 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_-_69129501 | 0.01 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr12_-_10978957 | 0.01 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr21_+_30673091 | 0.01 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr12_-_102224704 | 0.01 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr6_-_18249971 | 0.01 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr4_-_152149033 | 0.01 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr14_+_24407940 | 0.01 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr9_+_35673853 | 0.01 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr6_-_161695074 | 0.01 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr5_+_139739772 | 0.01 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr13_+_24144796 | 0.01 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chrX_+_135279179 | 0.01 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr2_+_170590321 | 0.01 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr2_+_29320571 | 0.01 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr17_+_28268623 | 0.01 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chrX_+_135278908 | 0.01 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr11_+_3968573 | 0.01 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr6_-_76782371 | 0.01 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr17_-_5321549 | 0.01 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr19_+_16940198 | 0.01 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr5_-_59481406 | 0.01 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr7_-_140482926 | 0.01 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr19_-_53696587 | 0.01 |
ENST00000396424.3
ENST00000600412.1 |
ZNF665
|
zinc finger protein 665 |
| chr11_+_98891797 | 0.01 |
ENST00000527185.1
ENST00000528682.1 ENST00000524871.1 |
CNTN5
|
contactin 5 |
| chr11_-_83393429 | 0.01 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr7_+_100026406 | 0.00 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr3_-_149470229 | 0.00 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr1_+_205197304 | 0.00 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr6_-_127840021 | 0.00 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr1_+_79115503 | 0.00 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr11_+_123430948 | 0.00 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr3_-_74570291 | 0.00 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr19_+_52873166 | 0.00 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr12_-_120241187 | 0.00 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr20_-_1447547 | 0.00 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr2_-_183387430 | 0.00 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_44281043 | 0.00 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr12_+_109785708 | 0.00 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |