Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
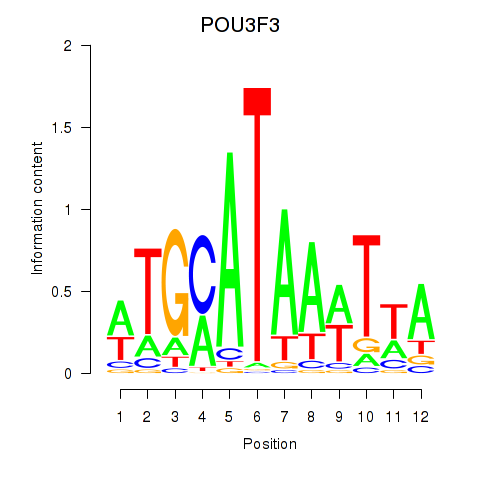
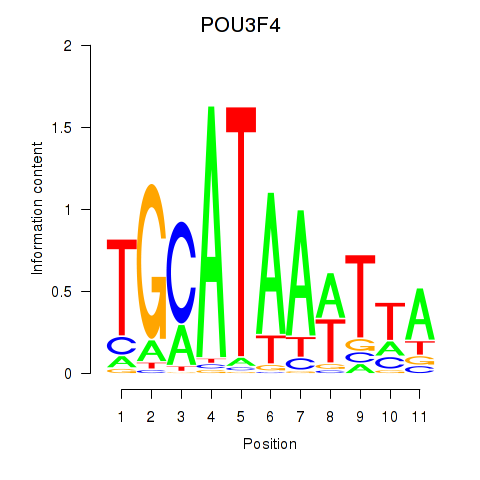
Results for POU3F3_POU3F4
Z-value: 0.83
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_45670717 | 0.77 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr6_+_144606817 | 0.75 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr15_+_76352178 | 0.74 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr1_+_81001398 | 0.62 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_-_151882031 | 0.60 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr15_-_45670924 | 0.59 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_+_81106951 | 0.55 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr8_-_25281747 | 0.50 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr21_+_17443521 | 0.37 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_-_146461027 | 0.37 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr15_-_76352069 | 0.37 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr7_-_100026280 | 0.36 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr2_-_101034070 | 0.33 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr12_-_42631529 | 0.33 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr6_+_148593425 | 0.31 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr14_+_21249200 | 0.29 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr2_+_217363559 | 0.29 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr6_-_10115007 | 0.29 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chrX_-_80457385 | 0.29 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr16_-_21314360 | 0.24 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr3_-_100558953 | 0.24 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_-_185826718 | 0.24 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr21_+_17443434 | 0.23 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_168155577 | 0.22 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_+_42756903 | 0.22 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr12_-_111926342 | 0.21 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr12_-_10324716 | 0.21 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_+_31496809 | 0.21 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr1_+_59775752 | 0.20 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr1_+_209602609 | 0.20 |
ENST00000458250.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chrM_+_5824 | 0.19 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr2_-_220264703 | 0.19 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr2_+_145780767 | 0.18 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr10_-_94003003 | 0.18 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_-_105332084 | 0.18 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr12_+_25205628 | 0.17 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_25205446 | 0.17 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_41221975 | 0.17 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr12_+_59989918 | 0.17 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_101702417 | 0.16 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr4_-_114438763 | 0.16 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr5_+_125759140 | 0.16 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr2_+_190541153 | 0.16 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr5_+_36608280 | 0.15 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_105010320 | 0.15 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr17_-_26220366 | 0.15 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr19_-_20748614 | 0.15 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr12_+_25205568 | 0.15 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr4_-_186570679 | 0.15 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_-_55208866 | 0.15 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr16_-_4164027 | 0.15 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chrX_-_119445306 | 0.14 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr5_-_42811986 | 0.14 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr18_-_52989217 | 0.14 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr17_+_66511540 | 0.14 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr2_+_166095898 | 0.14 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr15_+_63569785 | 0.14 |
ENST00000380343.4
ENST00000560353.1 |
APH1B
|
APH1B gamma secretase subunit |
| chr3_-_141747950 | 0.14 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr1_-_238108575 | 0.14 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr14_-_21270995 | 0.13 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_-_141747439 | 0.13 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_+_31497271 | 0.13 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr2_+_161993465 | 0.13 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr8_+_61591337 | 0.13 |
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr7_+_80267973 | 0.12 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_161035655 | 0.12 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr16_+_75690093 | 0.12 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr18_-_25739260 | 0.12 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr7_+_143771275 | 0.12 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chrX_+_28605516 | 0.12 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr4_-_110723335 | 0.12 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr7_-_229557 | 0.12 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr17_+_17206635 | 0.11 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr12_+_25205666 | 0.11 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr3_-_149375783 | 0.11 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr3_+_177545563 | 0.11 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr4_-_110723194 | 0.11 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr22_+_39101728 | 0.11 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr6_+_127898312 | 0.11 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_+_135596106 | 0.11 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_-_160279207 | 0.11 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr6_-_154751629 | 0.10 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr4_-_110723134 | 0.10 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr5_+_140753444 | 0.10 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chrX_+_80457442 | 0.10 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr1_+_61547405 | 0.10 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr12_-_25801478 | 0.10 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr2_+_135596180 | 0.10 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_+_158787098 | 0.10 |
ENST00000397832.2
ENST00000451172.1 ENST00000482126.1 |
IQCJ
|
IQ motif containing J |
| chr2_-_224702740 | 0.10 |
ENST00000444408.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr4_-_144940477 | 0.10 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr2_-_204398141 | 0.10 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr4_-_100575781 | 0.10 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr19_-_39402798 | 0.10 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr12_+_6554021 | 0.09 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr9_-_14180778 | 0.09 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr14_-_25479811 | 0.09 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr5_+_140762268 | 0.09 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr3_-_196242233 | 0.09 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr17_+_7387677 | 0.09 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr1_+_104198377 | 0.09 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr13_-_26795840 | 0.09 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr18_-_52989525 | 0.09 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr10_+_35484793 | 0.09 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr15_-_54267147 | 0.09 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr5_-_42812143 | 0.08 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_-_18632861 | 0.08 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr1_+_78383813 | 0.08 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr4_+_186990298 | 0.08 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr3_+_153202284 | 0.08 |
ENST00000446603.2
|
C3orf79
|
chromosome 3 open reading frame 79 |
| chr11_-_2906979 | 0.08 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr22_+_40322595 | 0.08 |
ENST00000420971.1
ENST00000544756.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr5_-_39462390 | 0.08 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_14880892 | 0.08 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr1_-_27701307 | 0.08 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr17_-_61996136 | 0.08 |
ENST00000342364.4
|
GH1
|
growth hormone 1 |
| chr6_-_49712147 | 0.08 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr13_-_46679144 | 0.08 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr6_+_143447322 | 0.08 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr6_-_76782371 | 0.08 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr8_-_124749609 | 0.08 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr12_+_49297887 | 0.08 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr2_-_46769694 | 0.08 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr4_+_30723003 | 0.08 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chrX_+_105936982 | 0.08 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr20_+_43990576 | 0.07 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr17_-_3599327 | 0.07 |
ENST00000551178.1
ENST00000552276.1 ENST00000547178.1 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr12_-_6960407 | 0.07 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr5_+_101569696 | 0.07 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr5_+_36608422 | 0.07 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_-_109935819 | 0.07 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr22_-_39268308 | 0.07 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_-_122107549 | 0.07 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr5_-_111312622 | 0.07 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr2_+_33359473 | 0.07 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr6_+_74171301 | 0.07 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr5_-_133510456 | 0.07 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr12_+_58003935 | 0.07 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr19_-_13900972 | 0.07 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr13_+_22245522 | 0.07 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr13_-_103389159 | 0.07 |
ENST00000322527.2
|
CCDC168
|
coiled-coil domain containing 168 |
| chr22_+_45148432 | 0.07 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr1_+_116915270 | 0.07 |
ENST00000418797.1
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr16_-_28192360 | 0.07 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr10_+_71561704 | 0.07 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_228244013 | 0.07 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr8_-_38008783 | 0.07 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr15_+_92006567 | 0.07 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr3_+_169539710 | 0.07 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr6_+_3982909 | 0.07 |
ENST00000356722.3
|
C6ORF50
|
C6ORF50 |
| chr15_+_49447947 | 0.07 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr6_-_32374900 | 0.07 |
ENST00000374995.3
ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2
|
butyrophilin-like 2 (MHC class II associated) |
| chr19_+_29493456 | 0.06 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr2_-_166930131 | 0.06 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr9_-_107367951 | 0.06 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chr7_-_38948774 | 0.06 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr2_+_112895939 | 0.06 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr19_+_107471 | 0.06 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr5_+_140705777 | 0.06 |
ENST00000606901.1
ENST00000606674.1 |
AC005618.6
|
AC005618.6 |
| chr17_-_61996160 | 0.06 |
ENST00000458650.2
ENST00000351388.4 ENST00000323322.5 |
GH1
|
growth hormone 1 |
| chrX_+_105855160 | 0.06 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr6_-_11779174 | 0.06 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr7_+_100026406 | 0.06 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr1_-_104239076 | 0.06 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr14_-_21270561 | 0.06 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr17_-_3599696 | 0.06 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr4_+_102711874 | 0.06 |
ENST00000508653.1
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr2_-_175711133 | 0.06 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr6_-_46459675 | 0.06 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr13_+_103459704 | 0.06 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr12_+_59989791 | 0.06 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_67142710 | 0.06 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr2_+_120687335 | 0.06 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr5_-_150080472 | 0.06 |
ENST00000521464.1
ENST00000518917.1 ENST00000447771.2 ENST00000540000.1 ENST00000199814.4 |
RBM22
|
RNA binding motif protein 22 |
| chr10_+_71561630 | 0.06 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr15_+_74422585 | 0.05 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_+_49297899 | 0.05 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr2_+_7017796 | 0.05 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr8_-_36636676 | 0.05 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr10_+_24738355 | 0.05 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr14_+_38033252 | 0.05 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr7_+_48211048 | 0.05 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr3_-_193272874 | 0.05 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr3_-_151047327 | 0.05 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr2_-_113993020 | 0.05 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr7_+_116593433 | 0.05 |
ENST00000323984.3
ENST00000393449.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr17_+_66511224 | 0.05 |
ENST00000588178.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr15_-_75249793 | 0.05 |
ENST00000322177.5
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr13_-_20110902 | 0.05 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr1_-_6614565 | 0.05 |
ENST00000377705.5
|
NOL9
|
nucleolar protein 9 |
| chr4_+_146403912 | 0.05 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr5_+_140474181 | 0.05 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr6_-_47277634 | 0.05 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr4_-_87281224 | 0.05 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_+_150065278 | 0.05 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr15_-_76304731 | 0.05 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr6_-_11779403 | 0.05 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_49712123 | 0.05 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.5 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.7 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:1901147 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.0 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 2.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |